KDM6B protects T-ALL cells from NOTCH1-induced oncogenic stress
- PMID: 36797416
- PMCID: PMC10081958
- DOI: 10.1038/s41375-023-01853-9
KDM6B protects T-ALL cells from NOTCH1-induced oncogenic stress
Abstract
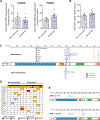
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematopoietic neoplasm resulting from the malignant transformation of T-cell progenitors. While activating NOTCH1 mutations are the dominant genetic drivers of T-ALL, epigenetic dysfunction plays a central role in the pathology of T-ALL and can provide alternative mechanisms to oncogenesis in lieu of or in combination with genetic mutations. The histone demethylase enzyme KDM6A (UTX) is also recurrently mutated in T-ALL patients and functions as a tumor suppressor. However, its gene paralog, KDM6B (JMJD3), is never mutated and can be significantly overexpressed, suggesting it may be necessary for sustaining the disease. Here, we used mouse and human T-ALL models to show that KDM6B is required for T-ALL development and maintenance. Using NOTCH1 gain-of-function retroviral models, mouse cells genetically deficient for Kdm6b were unable to propagate T-ALL. Inactivating KDM6B in human T-ALL patient cells by CRISPR/Cas9 showed KDM6B-targeted cells were significantly outcompeted over time. The dependence of T-ALL cells on KDM6B was proportional to the oncogenic strength of NOTCH1 mutation, with KDM6B required to prevent stress-induced apoptosis from strong NOTCH1 signaling. These studies identify a crucial role for KDM6B in sustaining NOTCH1-driven T-ALL and implicate KDM6B as a novel therapeutic target in these patients.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
DECLARATION OF INTERESTS
G.A.C. has performed consulting and received research support from Incyte (unrelated to this work). The remaining authors have declared that no conflict of interest exists.
Figures


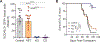



References
-
- Pui CH, Robison LL, and Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371(9617):1030–43. - PubMed
-
- Uckun FM, Gaynon PS, Sensel MG, Nachman J, Trigg ME, Steinherz PG, et al. Clinical features and treatment outcome of childhood T-lineage acute lymphoblastic leukemia according to the apparent maturational stage of T-lineage leukemic blasts: a Children’s Cancer Group study. J Clin Oncol. 1997;15(6):2214–21. - PubMed
-
- Goldberg JM, Silverman LB, Levy DE, Dalton VK, Gelber RD, Lehmann L, et al. Childhood T-cell acute lymphoblastic leukemia: the Dana-Farber Cancer Institute acute lymphoblastic leukemia consortium experience. J Clin Oncol. 2003;21(19):3616–22. - PubMed
-
- Oudot C, Auclerc MF, Levy V, Porcher R, Piguet C, Perel Y, et al. Prognostic factors for leukemic induction failure in children with acute lymphoblastic leukemia and outcome after salvage therapy: the FRALLE 93 study. J Clin Oncol. 2008;26(9):1496–503. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

