Efficient in vivo genome editing prevents hypertrophic cardiomyopathy in mice
- PMID: 36797483
- PMCID: PMC9941048
- DOI: 10.1038/s41591-022-02190-7
Efficient in vivo genome editing prevents hypertrophic cardiomyopathy in mice
Abstract
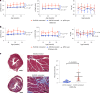
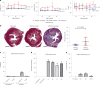
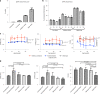
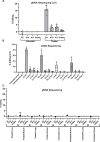
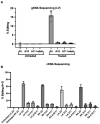
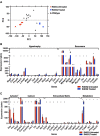
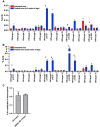
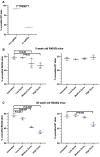
Dominant missense pathogenic variants in cardiac myosin heavy chain cause hypertrophic cardiomyopathy (HCM), a currently incurable disorder that increases risk for stroke, heart failure and sudden cardiac death. In this study, we assessed two different genetic therapies-an adenine base editor (ABE8e) and a potent Cas9 nuclease delivered by AAV9-to prevent disease in mice carrying the heterozygous HCM pathogenic variant myosin R403Q. One dose of dual-AAV9 vectors, each carrying one half of RNA-guided ABE8e, corrected the pathogenic variant in ≥70% of ventricular cardiomyocytes and maintained durable, normal cardiac structure and function. An additional dose provided more editing in the atria but also increased bystander editing. AAV9 delivery of RNA-guided Cas9 nuclease effectively inactivated the pathogenic allele, albeit with dose-dependent toxicities, necessitating a narrow therapeutic window to maintain health. These preclinical studies demonstrate considerable potential for single-dose genetic therapies to correct or silence pathogenic variants and prevent the development of HCM.
© 2023. The Author(s).
Conflict of interest statement
L.H.V. holds equity and serves on the Board of Directors for Affinia Therapeutics and Ciendias Bio, where he is employed, and holds equity in Akouos. He has licensed technology to Affinia, Akouos and Novartis. D.R.L. is a consultant and equity owner of Beam Therapeutics, Prime Medicine, Pairwise Plants and Chroma Medicine, companies that use genome editing or genome engineering. J.G.S. and C.E.S are founders of Myokardia (a Bristol Myers Squibb subsidiary) and are consultants for Maze and BridgeBio. C.E.S serves on the Merck Board of Directors and the Burroughs Wellcome Fund. None of these companies had any input into the design, execution, analyses or writing of this manuscript. Other authors declare no competing interests.
Figures






Comment in
-
CRISPR gene-editing therapies for hypertrophic cardiomyopathy.Nat Med. 2023 Feb;29(2):305-306. doi: 10.1038/s41591-022-02184-5. Nat Med. 2023. PMID: 36797479 Free PMC article.
-
Genome editing prevents hypertrophic cardiomyopathy in mice.Nat Rev Cardiol. 2023 Apr;20(4):211. doi: 10.1038/s41569-023-00852-8. Nat Rev Cardiol. 2023. PMID: 36849814 No abstract available.
-
Testing the genome-editing toolkit in cardiomyopathy.Nat Rev Drug Discov. 2023 Apr;22(4):270. doi: 10.1038/d41573-023-00046-4. Nat Rev Drug Discov. 2023. PMID: 36899271 No abstract available.
-
CRISPRing the hypertrophic cardiomyopathy: correcting one pathogenic variant at a time.Signal Transduct Target Ther. 2023 Jun 26;8(1):254. doi: 10.1038/s41392-023-01526-0. Signal Transduct Target Ther. 2023. PMID: 37365168 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

