Analysis of genetically determined gene expression suggests role of inflammatory processes in exfoliation syndrome
- PMID: 36797672
- PMCID: PMC9936777
- DOI: 10.1186/s12864-023-09179-7
Analysis of genetically determined gene expression suggests role of inflammatory processes in exfoliation syndrome
Abstract
Background: Exfoliation syndrome (XFS) is an age-related systemic disorder characterized by excessive production and progressive accumulation of abnormal extracellular material, with pathognomonic ocular manifestations. It is the most common cause of secondary glaucoma, resulting in widespread global blindness. The largest global meta-analysis of XFS in 123,457 multi-ethnic individuals from 24 countries identified seven loci with the strongest association signal in chr15q22-25 region near LOXL1. Expression analysis have so far correlated coding and a few non-coding variants in the region with LOXL1 expression levels, but functional effects of these variants is unclear. We hypothesize that analysis of the contribution of the genetically determined component of gene expression to XFS risk can provide a powerful method to elucidate potential roles of additional genes and clarify biology that underlie XFS.
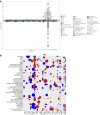
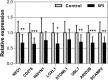
Results: Transcriptomic Wide Association Studies (TWAS) using PrediXcan models trained in 48 GTEx tissues leveraging on results from the multi-ethnic and European ancestry GWAS were performed. To eliminate the possibility of false-positive results due to Linkage Disequilibrium (LD) contamination, we i) performed PrediXcan analysis in reduced models removing variants in LD with LOXL1 missense variants associated with XFS, and variants in LOXL1 models in both multiethnic and European ancestry individuals, ii) conducted conditional analysis of the significant signals in European ancestry individuals, and iii) filtered signals based on correlated gene expression, LD and shared eQTLs, iv) conducted expression validation analysis in human iris tissues. We observed twenty-eight genes in chr15q22-25 region that showed statistically significant associations, which were whittled down to ten genes after statistical validations. In experimental analysis, mRNA transcript levels for ARID3B, CD276, LOXL1, NEO1, SCAMP2, and UBL7 were significantly decreased in iris tissues from XFS patients compared to control samples. TWAS genes for XFS were significantly enriched for genes associated with inflammatory conditions. We also observed a higher incidence of XFS comorbidity with inflammatory and connective tissue diseases.
Conclusion: Our results implicate a role for connective tissues and inflammation pathways in the etiology of XFS. Targeting the inflammatory pathway may be a potential therapeutic option to reduce progression in XFS.
Keywords: Exfoliation syndrome; GTEx; GWAS; TWAS; predicted expressions; transcriptomics.
© 2023. The Author(s).
Conflict of interest statement
ERG receives an honorarium from the journal
The remaining authors declare no competing interests.
Figures

Similar articles
-
Genetic association study of exfoliation syndrome identifies a protective rare variant at LOXL1 and five new susceptibility loci.Nat Genet. 2017 Jul;49(7):993-1004. doi: 10.1038/ng.3875. Epub 2017 May 29. Nat Genet. 2017. PMID: 28553957 Free PMC article.
-
Expression and regulation of LOXL1 and elastin-related genes in eyes with exfoliation syndrome.J Glaucoma. 2014 Oct-Nov;23(8 Suppl 1):S48-50. doi: 10.1097/IJG.0000000000000120. J Glaucoma. 2014. PMID: 25275906 Review.
-
Genetic variants and cellular stressors associated with exfoliation syndrome modulate promoter activity of a lncRNA within the LOXL1 locus.Hum Mol Genet. 2015 Nov 15;24(22):6552-63. doi: 10.1093/hmg/ddv347. Epub 2015 Aug 25. Hum Mol Genet. 2015. PMID: 26307087 Free PMC article.
-
LOXL1 gene polymorphisms are associated with exfoliation syndrome/exfoliation glaucoma risk: An updated meta-analysis.PLoS One. 2021 Apr 28;16(4):e0250772. doi: 10.1371/journal.pone.0250772. eCollection 2021. PLoS One. 2021. PMID: 33909695 Free PMC article.
-
From epidemiology to lysyl oxidase like one (LOXL1) polymorphisms discovery: phenotyping and genotyping exfoliation syndrome and exfoliation glaucoma in Iceland.Acta Ophthalmol. 2009 Aug;87(5):478-87. doi: 10.1111/j.1755-3768.2009.01635.x. Acta Ophthalmol. 2009. PMID: 19664108 Review.
Cited by
-
Hematological inflammation biomarkers in patients with pseudoexfoliation syndrome and pseudoexfoliation glaucoma.Int J Ophthalmol. 2025 Jun 18;18(6):1042-1052. doi: 10.18240/ijo.2025.06.10. eCollection 2025. Int J Ophthalmol. 2025. PMID: 40534792 Free PMC article.
-
Comprehensive Proteomic Profiling of Exfoliation Glaucoma Via Mass Spectrometry Reveals SVEP1 as a Potential Biomarker.Invest Ophthalmol Vis Sci. 2025 Mar 3;66(3):19. doi: 10.1167/iovs.66.3.19. Invest Ophthalmol Vis Sci. 2025. PMID: 40052860 Free PMC article.
-
Association of Inflammatory and Ischemic Markers with Posterior Segment Parameters in Pseudoexfoliation Syndrome and Glaucoma.J Clin Med. 2025 May 29;14(11):3833. doi: 10.3390/jcm14113833. J Clin Med. 2025. PMID: 40507594 Free PMC article.
-
Structural framework to address variant-gene relationship in primary open-angle glaucoma.Vision Res. 2025 Jan;226:108505. doi: 10.1016/j.visres.2024.108505. Epub 2024 Nov 8. Vision Res. 2025. PMID: 39520803 Review.
-
Exfoliation syndrome genetics in the era of post-GWAS.Vision Res. 2025 Jan;226:108518. doi: 10.1016/j.visres.2024.108518. Epub 2024 Nov 16. Vision Res. 2025. PMID: 39549468 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

