Therapeutic immunomodulation by rationally designed nucleic acids and nucleic acid nanoparticles
- PMID: 36798121
- PMCID: PMC9927404
- DOI: 10.3389/fimmu.2023.1053550
Therapeutic immunomodulation by rationally designed nucleic acids and nucleic acid nanoparticles
Abstract
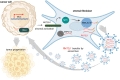
The immune system has evolved to defend organisms against exogenous threats such as viruses, bacteria, fungi, and parasites by distinguishing between "self" and "non-self". In addition, it guards us against other diseases, such as cancer, by detecting and responding to transformed and senescent cells. However, for survival and propagation, the altered cells and invading pathogens often employ a wide range of mechanisms to avoid, inhibit, or manipulate the immunorecognition. As such, the development of new modes of therapeutic intervention to augment protective and prevent harmful immune responses is desirable. Nucleic acids are biopolymers essential for all forms of life and, therefore, delineating the complex defensive mechanisms developed against non-self nucleic acids can offer an exciting avenue for future biomedicine. Nucleic acid technologies have already established numerous approaches in therapy and biotechnology; recently, rationally designed nucleic acids nanoparticles (NANPs) with regulated physiochemical properties and biological activities has expanded our repertoire of therapeutic options. When compared to conventional therapeutic nucleic acids (TNAs), NANP technologies can be rendered more beneficial for synchronized delivery of multiple TNAs with defined stabilities, immunological profiles, and therapeutic functions. This review highlights several recent advances and possible future directions of TNA and NANP technologies that are under development for controlled immunomodulation.
Keywords: DAMP; NANPs; PAMP; PRR; immunomodulation; innate immune system; therapy.
Copyright © 2023 Panigaj, Skelly, Beasock, Marriott, Johnson, Salotti and Afonin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Structure and Composition Define Immunorecognition of Nucleic Acid Nanoparticles.Nano Lett. 2018 Jul 11;18(7):4309-4321. doi: 10.1021/acs.nanolett.8b01283. Epub 2018 Jun 20. Nano Lett. 2018. PMID: 29894623 Free PMC article.
-
Nucleic acid nanoparticles (NANPs) as molecular tools to direct desirable and avoid undesirable immunological effects.Adv Drug Deliv Rev. 2021 Jun;173:427-438. doi: 10.1016/j.addr.2021.04.011. Epub 2021 Apr 20. Adv Drug Deliv Rev. 2021. PMID: 33857556 Free PMC article. Review.
-
A cationic amphiphilic co-polymer as a carrier of nucleic acid nanoparticles (Nanps) for controlled gene silencing, immunostimulation, and biodistribution.Nanomedicine. 2020 Jan;23:102094. doi: 10.1016/j.nano.2019.102094. Epub 2019 Oct 25. Nanomedicine. 2020. PMID: 31669854 Free PMC article.
-
Exosomes as natural delivery carriers for programmable therapeutic nucleic acid nanoparticles (NANPs).Adv Drug Deliv Rev. 2021 Sep;176:113835. doi: 10.1016/j.addr.2021.113835. Epub 2021 Jun 16. Adv Drug Deliv Rev. 2021. PMID: 34144087 Free PMC article. Review.
-
Exosome mediated delivery of functional nucleic acid nanoparticles (NANPs).Nanomedicine. 2020 Nov;30:102285. doi: 10.1016/j.nano.2020.102285. Epub 2020 Aug 8. Nanomedicine. 2020. PMID: 32781137 Free PMC article.
Cited by
-
The scientific journey of a novel adjuvant (AS37) from bench to bedside.NPJ Vaccines. 2024 Feb 8;9(1):26. doi: 10.1038/s41541-024-00810-6. NPJ Vaccines. 2024. PMID: 38332005 Free PMC article. Review.
-
Immunostimulation of Fibrous Nucleic Acid Nanoparticles Can be Modulated through Aptamer-Based Functional Moieties: Unveiling the Structure-Activity Relationship and Mechanistic Insights.ACS Appl Mater Interfaces. 2024 Feb 21;16(7):8430-8441. doi: 10.1021/acsami.3c17779. Epub 2024 Feb 12. ACS Appl Mater Interfaces. 2024. PMID: 38344840 Free PMC article.
-
Autonomous Nucleic Acid and Protein Nanocomputing Agents Engineered to Operate in Living Cells.ACS Nano. 2025 Jan 21;19(2):1865-1883. doi: 10.1021/acsnano.4c13663. Epub 2025 Jan 6. ACS Nano. 2025. PMID: 39760461 Free PMC article. Review.
-
Cracking the Code: Enhancing Molecular Tools for Progress in Nanobiotechnology.ACS Appl Bio Mater. 2024 Jun 17;7(6):3587-3604. doi: 10.1021/acsabm.4c00432. Epub 2024 Jun 4. ACS Appl Bio Mater. 2024. PMID: 38833534 Free PMC article. Review.
-
Nucleic acid drugs: recent progress and future perspectives.Signal Transduct Target Ther. 2024 Nov 29;9(1):316. doi: 10.1038/s41392-024-02035-4. Signal Transduct Target Ther. 2024. PMID: 39609384 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

