This is a preprint.
A Potent and Selective CDKL5/GSK3 Chemical Probe is Neuroprotective
- PMID: 36798313
- PMCID: PMC9934649
- DOI: 10.1101/2023.02.09.527935
A Potent and Selective CDKL5/GSK3 Chemical Probe is Neuroprotective
Update in
-
Discovery of a Potent and Selective CDKL5/GSK3 Chemical Probe That Is Neuroprotective.ACS Chem Neurosci. 2023 May 3;14(9):1672-1685. doi: 10.1021/acschemneuro.3c00135. Epub 2023 Apr 21. ACS Chem Neurosci. 2023. PMID: 37084253 Free PMC article.
Abstract
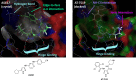
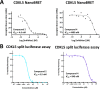
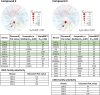
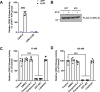
Despite mediating several essential processes in the brain, including during development, cyclin-dependent kinase-like 5 (CDKL5) remains a poorly characterized human protein kinase. Accordingly, its substrates, functions, and regulatory mechanisms have not been fully described. We realized that availability of a potent and selective small molecule probe targeting CDKL5 could enable illumination of its roles in normal development as well as in diseases where it has become aberrant due to mutation. We prepared analogs of AT-7519, a known inhibitor of several cyclin dependent and cyclin-dependent kinase-like kinases that has been advanced into Phase II clinical trials. We identified analog 2 as a highly potent and cell-active chemical probe for CDKL5/GSK3 (glycogen synthase kinase 3). Evaluation of its kinome-wide selectivity confirmed that analog 2 demonstrates excellent selectivity and only retains GSK3α/β affinity. As confirmation that our chemical probe is a high-quality tool to use in directed biological studies, we demonstrated inhibition of downstream CDKL5 and GSK3α/β signaling and solved a co-crystal structure of analog 2 bound to CDKL5. A structurally similar analog ( 4 ) proved to lack CDKL5 affinity and maintain potent and selective inhibition of GSK3α/β. Finally, we used our chemical probe pair ( 2 and 4 ) to demonstrate that inhibition of CDKL5 and/or GSK3α/β promotes the survival of human motor neurons exposed to endoplasmic reticulum (ER) stress. We have demonstrated a neuroprotective phenotype elicited by our chemical probe pair and exemplified the utility of our compounds to characterize the role of CDKL5/GSK3 in neurons and beyond.
Conflict of interest statement
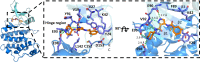
The authors declare no competing financial interests. The crystallographic coordinates of the CDKL5 co-structure have been deposited in the Protein Data Bank as 8CIE.
Figures
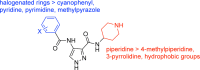



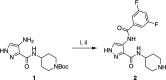
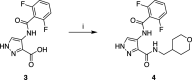
References
-
- Ricciardi S.; Ungaro F.; Hambrock M.; Rademacher N.; Stefanelli G.; Brambilla D.; Sessa A.; Magagnotti C.; Bachi A.; Giarda E.; et al. CDKL5 ensures excitatory synapse stability by reinforcing NGL-1-PSD95 interaction in the postsynaptic compartment and is impaired in patient iPSC-derived neurons. Nat Cell Biol 2012, 14 (9), 911–923. DOI: 10.1038/ncb2566. - DOI - PMC - PubMed
-
- Fuchs C.; Trazzi S.; Torricella R.; Viggiano R.; De Franceschi M.; Amendola E.; Gross C.; Calzà L.; Bartesaghi R.; Ciani E. Loss of CDKL5 impairs survival and dendritic growth of newborn neurons by altering AKT/GSK-3β signaling. Neurobiol Dis 2014, 70 (100), 53–68. DOI: 10.1016/j.nbd.2014.06.006. - DOI - PMC - PubMed
-
- Barbiero I.; Valente D.; Chandola C.; Magi F.; Bergo A.; Monteonofrio L.; Tramarin M.; Fazzari M.; Soddu S.; Landsberger N.; et al. CDKL5 localizes at the centrosome and midbody and is required for faithful cell division. Sci Rep 2017, 7 (1), 6228. DOI: 10.1038/s41598-017-05875-z. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
