Distinct dosage compensations of ploidy-sensitive and -insensitive X chromosome genes during development and in diseases
- PMID: 36798435
- PMCID: PMC9926305
- DOI: 10.1016/j.isci.2023.105997
Distinct dosage compensations of ploidy-sensitive and -insensitive X chromosome genes during development and in diseases
Abstract
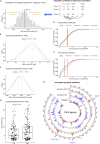
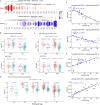
The active X chromosome in mammals is upregulated to balance its dosage to autosomes during evolution. However, it is elusive why the known dosage compensation machinery showed uneven and small influence on X genes. Here, based on >20,000 transcriptomes, we identified two X gene groups (ploidy-sensitive [PSX] and ploidy-insensitive [PIX]), showing distinct but evolutionarily conserved dosage compensations (termed XAR). We demonstrated that XAR-PIX was downregulated whereas XAR-PSX upregulated at both RNA and protein levels across cancer types, in contrast with their trends during stem cell differentiation. XAR-PIX, but not XAR-PSX, was lower and correlated with autoantibodies and inflammation in patients of lupus, suggesting that insufficient dosage of PIX genes contribute to lupus pathogenesis. We further identified and experimentally validated two XAR regulators, TP53 and ATRX. Collectively, we provided insights into X dosage compensation in mammals and demonstrated different regulation of PSX and PIX and their pathophysiological roles in human diseases.
Keywords: Biological sciences; Genetics; Molecular physiology.
© 2023 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Deng X., Berletch J.B., Ma W., Nguyen D.K., Hiatt J.B., Noble W.S., Shendure J., Disteche C.M. Mammalian X upregulation is associated with enhanced transcription initiation, RNA half-life, and MOF-mediated H4K16 acetylation. Dev. Cell. 2013;25:55–68. doi: 10.1016/j.devcel.2013.01.028. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

