FACILITATE: A real-world, multicenter, prospective study investigating the utility of a rapid, fully automated real-time PCR assay versus local reference methods for detecting epidermal growth factor receptor variants in NSCLC
- PMID: 36798672
- PMCID: PMC9927408
- DOI: 10.3389/pore.2023.1610707
FACILITATE: A real-world, multicenter, prospective study investigating the utility of a rapid, fully automated real-time PCR assay versus local reference methods for detecting epidermal growth factor receptor variants in NSCLC
Abstract
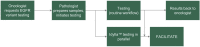
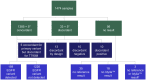
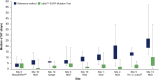
Accurate testing for epidermal growth factor receptor (EGFR) variants is essential for informing treatment decisions in non-small cell lung cancer (NSCLC). Automated diagnostic workflows may allow more streamlined initiation of targeted treatments, where appropriate, while comprehensive variant analysis is ongoing. FACILITATE, a real-world, prospective, multicenter, European study, evaluated performance and analytical turnaround time of the Idylla™ EGFR Mutation Test compared with local reference methods. Sixteen sites obtained formalin-fixed paraffin-embedded biopsy samples with ≥ 10% neoplastic cells from patients with NSCLC. Consecutive 5 μm sections from patient samples were tested for clinically relevant NSCLC-associated EGFR variants using the Idylla™ EGFR Mutation Test and local reference methods; performance (concordance) and analytical turnaround time were compared. Between January 2019 and November 2020, 1,474 parallel analyses were conducted. Overall percentage agreement was 97.7% [n = 1,418; 95% confidence interval (CI): 96.8-98.3], positive agreement, 87.4% (n = 182; 95% CI: 81.8-91.4) and negative agreement, 99.2% (n = 1,236; 95% CI: 98.5-99.6). There were 38 (2.6%) discordant cases. Ninety percent of results were returned with an analytical turnaround time of within 1 week using the Idylla™ EGFR Mutation Test versus ∼22 days using reference methods. The Idylla™ EGFR Mutation Test performed well versus local methods and had shorter analytical turnaround time. The Idylla™ EGFR Mutation Test can thus support application of personalized medicine in NSCLC.
Keywords: DNA mutational analysis; clinical decision-making; epidermal growth factor receptor; non-small cell lung cancer; turnaround time.
Copyright © 2023 Behnke, Cayre, De Maglio, Giannini, Habran, Tarsitano, Chetta, Cappellen, Lespagnol, Le Naoures, Massazza, Destro, Bonzheim, Rau, Battmann, Kah, Watkin and Hummel.
Conflict of interest statement
IB declares receipt of honoraria from Novartis, Bayer, Pfizer, Takeda, AstraZeneca and BMS. EW declares the receipt of honoraria from AstraZeneca and MSD. MH declares membership in advisory councils or committees for AstraZeneca, Roche, Novartis, Pierre Fabre GDM, Sanofi, MSD and BMS; and receipt of grants or funds from AstraZeneca. Author EW was employed by the company CYPATH. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The authors declare that this study received funding from AstraZeneca, Cambridge, UK and Biocartis, Mechelen, Belgium. Biocartis were involved in study design, and analysis and interpretation of data. AstraZeneca were not involved in study design or analysis and interpretation of data. The funders were not involved in collection of data. The funders reviewed the manuscript before submission.
Figures

References
-
- Planchard D, Popat S, Kerr K, Novello S, Smit EF, Faivre-Finn C, et al. Metastatic non-small cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Updated version published 15 september 2020 by the ESMO guidelines committee [online]. (2020) (2020). Available: https://www.esmo.org/guidelines/lung-and-chest-tumours/clinical-practice... (Accessed September 25, 2020).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

