Evidence Supporting That C-to-U RNA Editing Is the Major Force That Drives SARS-CoV-2 Evolution
- PMID: 36799984
- PMCID: PMC9936484
- DOI: 10.1007/s00239-023-10097-1
Evidence Supporting That C-to-U RNA Editing Is the Major Force That Drives SARS-CoV-2 Evolution
Abstract
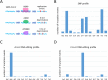
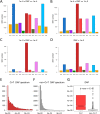
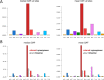
Mutations of DNA organisms are introduced by replication errors. However, SARS-CoV-2, as an RNA virus, is additionally subjected to rampant RNA editing by hosts. Both resources contributed to SARS-CoV-2 mutation and evolution, but the relative prevalence of the two origins is unknown. We performed comparative genomic analyses at intra-species (world-wide SARS-CoV-2 strains) and inter-species (SARS-CoV-2 and RaTG13 divergence) levels. We made prior predictions of the proportion of each mutation type (nucleotide substitution) under different scenarios and compared the observed versus the expected. C-to-T alteration, representing C-to-U editing, is far more abundant that all other mutation types. Derived allele frequency (DAF) as well as novel mutation rate of C-to-T are the highest in SARS-CoV-2 population, and C-T substitution dominates the divergence sites between SARS-CoV-2 and RaTG13. This is compelling evidence suggesting that C-to-U RNA editing is the major source of SARS-CoV-2 mutation. While replication errors serve as a baseline of novel mutation rate, the C-to-U editing has elevated the mutation rate for orders of magnitudes and accelerates the evolution of the virus.
Keywords: Allele frequency; C-to-U RNA editing; Evolution; Mutation; SARS-CoV-2.
© 2023. The Author(s), under exclusive licence to Springer Science+Business Media, LLC, part of Springer Nature.
Conflict of interest statement
The authors declare they have no conflict of interest.
Figures




References
-
- Byng JW, Chase MW, Christenhusz MJM, Fay MF, Judd WS, Mabberley DJ, Sennikov AN, Soltis DE, Soltis PS, Stevens PF, et al. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 2016;181:1–20. doi: 10.1111/boj.12385. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

