IL-21R signal reprogramming cooperates with CD40 and BCR signals to select and differentiate germinal center B cells
- PMID: 36800413
- PMCID: PMC10206726
- DOI: 10.1126/sciimmunol.add1823
IL-21R signal reprogramming cooperates with CD40 and BCR signals to select and differentiate germinal center B cells
Abstract
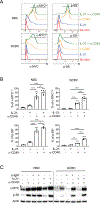
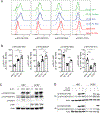
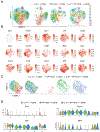
Both B cell receptor (BCR) and CD40 signaling are rewired in germinal center (GC) B cells (GCBCs) to synergistically induce c-MYC and phosphorylated S6 ribosomal protein (p-S6), markers of positive selection. How interleukin-21 (IL-21), a key T follicular helper (TFH)-derived cytokine, affects GCBCs is unclear. Like BCR and CD40 signals, IL-21 receptor (IL-21R) plus CD40 signals also synergize to induce c-MYC and p-S6 in GCBCs. However, IL-21R plus CD40 stimulation differentially affects GCBC fate compared with BCR plus CD40 ligation-engaging unique molecular mechanisms-as revealed by bulk RNA sequencing (RNA-seq), single-cell RNA-seq, and flow cytometry of GCBCs in vitro and in vivo. Whereas both signal pairs induced BLIMP1 in some GCBCs, only the IL-21R/CD40 combination induced IRF4hi/CD138+ cells, indicative of plasma cell differentiation, along with CCR6+/CD38+ memory B cell precursors. These findings reveal a second positive selection pathway in GCBCs, document rewired IL-21R signaling in GCBCs, and link specific TFH- and Ag-derived signals to GCBC differentiation.
Conflict of interest statement
Figures




Similar articles
-
The AKT kinase signaling network is rewired by PTEN to control proximal BCR signaling in germinal center B cells.Nat Immunol. 2019 Jun;20(6):736-746. doi: 10.1038/s41590-019-0376-3. Epub 2019 Apr 22. Nat Immunol. 2019. PMID: 31011187 Free PMC article.
-
CD40 and B cell antigen receptor dual triggering of resting B lymphocytes turns on a partial germinal center phenotype.J Exp Med. 1996 Jan 1;183(1):77-85. doi: 10.1084/jem.183.1.77. J Exp Med. 1996. PMID: 8551247 Free PMC article.
-
B Cell Receptor and CD40 Signaling Are Rewired for Synergistic Induction of the c-Myc Transcription Factor in Germinal Center B Cells.Immunity. 2018 Feb 20;48(2):313-326.e5. doi: 10.1016/j.immuni.2018.01.008. Immunity. 2018. PMID: 29396161 Free PMC article.
-
Linking signaling and selection in the germinal center.Immunol Rev. 2019 Mar;288(1):49-63. doi: 10.1111/imr.12744. Immunol Rev. 2019. PMID: 30874353 Free PMC article. Review.
-
The unusual metabolism of germinal center B cells.Trends Immunol. 2025 May;46(5):416-428. doi: 10.1016/j.it.2025.02.015. Epub 2025 Apr 12. Trends Immunol. 2025. PMID: 40221291 Review.
Cited by
-
Temporal dynamics and genomic programming of plasma cell fates.Nat Immunol. 2024 Jun;25(6):1097-1109. doi: 10.1038/s41590-024-01831-y. Epub 2024 May 2. Nat Immunol. 2024. PMID: 38698087
-
Immune Checkpoints in B Cells: Unlocking New Potentials in Cancer Treatment.Adv Sci (Weinh). 2024 Dec;11(47):e2403423. doi: 10.1002/advs.202403423. Epub 2024 Nov 7. Adv Sci (Weinh). 2024. PMID: 39509319 Free PMC article. Review.
-
CD4 T Cell-Derived IL-21 Is Critical for Sustaining Plasmodium Infection-Induced Germinal Center Responses and Promoting the Selection of Memory B Cells with Recall Potential.J Immunol. 2024 May 1;212(9):1467-1478. doi: 10.4049/jimmunol.2300683. J Immunol. 2024. PMID: 38477614 Free PMC article.
-
Accumulation of immune-suppressive CD4 + T cells in aging - tempering inflammaging at the expense of immunity.Semin Immunol. 2023 Nov;70:101836. doi: 10.1016/j.smim.2023.101836. Epub 2023 Aug 24. Semin Immunol. 2023. PMID: 37632992 Free PMC article. Review.
-
Imbalance of T follicular helper cell subsets trigger the differentiation of pathogenic B cells in idiopathic membranous nephropathy.Inflamm Res. 2024 Apr;73(4):485-498. doi: 10.1007/s00011-023-01838-5. Epub 2024 Mar 11. Inflamm Res. 2024. PMID: 38467875
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

