Artificial intelligence-based clustering and characterization of Parkinson's disease trajectories
- PMID: 36801900
- PMCID: PMC9938890
- DOI: 10.1038/s41598-023-30038-8
Artificial intelligence-based clustering and characterization of Parkinson's disease trajectories
Abstract
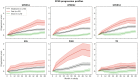
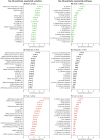
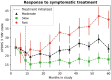
Parkinson's disease (PD) is a highly heterogeneous disease both with respect to arising symptoms and its progression over time. This hampers the design of disease modifying trials for PD as treatments which would potentially show efficacy in specific patient subgroups could be considered ineffective in a heterogeneous trial cohort. Establishing clusters of PD patients based on their progression patterns could help to disentangle the exhibited heterogeneity, highlight clinical differences among patient subgroups, and identify the biological pathways and molecular players which underlie the evident differences. Further, stratification of patients into clusters with distinct progression patterns could help to recruit more homogeneous trial cohorts. In the present work, we applied an artificial intelligence-based algorithm to model and cluster longitudinal PD progression trajectories from the Parkinson's Progression Markers Initiative. Using a combination of six clinical outcome scores covering both motor and non-motor symptoms, we were able to identify specific clusters of PD that showed significantly different patterns of PD progression. The inclusion of genetic variants and biomarker data allowed us to associate the established progression clusters with distinct biological mechanisms, such as perturbations in vesicle transport or neuroprotection. Furthermore, we found that patients of identified progression clusters showed significant differences in their responsiveness to symptomatic treatment. Taken together, our work contributes to a better understanding of the heterogeneity encountered when examining and treating patients with PD, and points towards potential biological pathways and genes that could underlie those differences.
© 2023. The Author(s).
Conflict of interest statement
PD and MA are employees of UCB BioPharma. HF, AAh, and AAv were full time employees of UCB BioPharma at the start of this study. NJM is a Veramed statistical consultant for UCB Biopharma.
Figures
References
-
- Sieber BA, Landis S, Koroshetz W, Bateman R, Siderowf A, Galpern WR, Parkinson's Disease 2014: Advancing Research, Improving Lives Conference Organizing Committee Prioritized research recommendations from the National Institute of Neurological Disorders and Stroke Parkinson's Disease 2014 conference. Annals of neurology. 2014;76(4):469–472. doi: 10.1002/ana.24261. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

