Deficiency of the minor spliceosome component U4atac snRNA secondarily results in ciliary defects in human and zebrafish
- PMID: 36802443
- PMCID: PMC9992838
- DOI: 10.1073/pnas.2102569120
Deficiency of the minor spliceosome component U4atac snRNA secondarily results in ciliary defects in human and zebrafish
Abstract
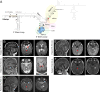
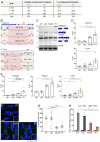
In the human genome, about 750 genes contain one intron excised by the minor spliceosome. This spliceosome comprises its own set of snRNAs, among which U4atac. Its noncoding gene, RNU4ATAC, has been found mutated in Taybi-Linder (TALS/microcephalic osteodysplastic primordial dwarfism type 1), Roifman (RFMN), and Lowry-Wood (LWS) syndromes. These rare developmental disorders, whose physiopathological mechanisms remain unsolved, associate ante- and post-natal growth retardation, microcephaly, skeletal dysplasia, intellectual disability, retinal dystrophy, and immunodeficiency. Here, we report bi-allelic RNU4ATAC mutations in five patients presenting with traits suggestive of the Joubert syndrome (JBTS), a well-characterized ciliopathy. These patients also present with traits typical of TALS/RFMN/LWS, thus widening the clinical spectrum of RNU4ATAC-associated disorders and indicating ciliary dysfunction as a mechanism downstream of minor splicing defects. Intriguingly, all five patients carry the n.16G>A mutation, in the Stem II domain, either at the homozygous or compound heterozygous state. A gene ontology term enrichment analysis on minor intron-containing genes reveals that the cilium assembly process is over-represented, with no less than 86 cilium-related genes containing at least one minor intron, among which there are 23 ciliopathy-related genes. The link between RNU4ATAC mutations and ciliopathy traits is supported by alterations of primary cilium function in TALS and JBTS-like patient fibroblasts, as well as by u4atac zebrafish model, which exhibits ciliopathy-related phenotypes and ciliary defects. These phenotypes could be rescued by WT but not by pathogenic variants-carrying human U4atac. Altogether, our data indicate that alteration of cilium biogenesis is part of the physiopathological mechanisms of TALS/RFMN/LWS, secondarily to defects of minor intron splicing.
Keywords: U4atac; genetic disease; minor introns; primary cilium; splicing.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- Edery P., et al. , Association of TALS developmental disorder with defect in minor splicing component U4atac snRNA. Science 332, 240–243 (2011). - PubMed
-
- Hagiwara H., et al. , Immunodeficiency in a patient with microcephalic osteodysplastic primordial dwarfism type I as compared to Roifman syndrome. Brain Dev. 43, 337–342 (2020). - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

