Structure and function of prodrug-activating peptidases
- PMID: 36803695
- PMCID: PMC10030199
- DOI: 10.1016/j.biochi.2022.07.019
Structure and function of prodrug-activating peptidases
Abstract
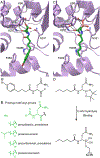
Bacteria protect themselves from the toxicity of antimicrobial metabolites they produce through several strategies. In one resistance mechanism, bacteria assemble a non-toxic precursor on an N-acyl-d-asparagine prodrug motif in the cytoplasm, then export it to the periplasm where a dedicated d-amino peptidase hydrolyzes the prodrug motif. These prodrug-activating peptidases contain an N-terminal periplasmic S12 hydrolase domain and C-terminal transmembrane domains (TMDs) of varying lengths: type I peptidases contain three transmembrane helices, and type II peptidases have an additional C-terminal ABC half-transporter. We review studies which have addressed the role of the TMD in function, the substrate specificity, and the biological assembly of ClbP, the type I peptidase that activates colibactin. We use modeling and sequence analyses to extend those insights to other prodrug-activating peptidases and ClbP-like proteins which are not part of prodrug resistance gene clusters. These ClbP-like proteins may play roles in the biosynthesis or degradation of other natural products, including antibiotics, may adopt different TMD folds, and have different substrate specificity compared to prodrug-activating homologs. Finally, we review the data supporting the long-standing hypothesis that ClbP interacts with transporters in the cell and that this association is important for the export of other natural products. Future investigations of this hypothesis as well as of the structure and function of type II peptidases will provide a complete account of the role of prodrug-activating peptidases in the activation and secretion of bacterial toxins.
Keywords: Bacterial toxin; Beta-lactamase; Membrane-embedded peptidase; Non-ribosomal peptide synthesis; Prodrug resistance mechanism; Sequence similarity network.
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
Figures


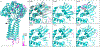
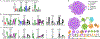

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

