Single-Cell Analysis Reveals Distinct Immune and Smooth Muscle Cell Populations that Contribute to Chronic Thromboembolic Pulmonary Hypertension
- PMID: 36803741
- PMCID: PMC10595445
- DOI: 10.1164/rccm.202203-0441OC
Single-Cell Analysis Reveals Distinct Immune and Smooth Muscle Cell Populations that Contribute to Chronic Thromboembolic Pulmonary Hypertension
Abstract
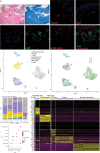
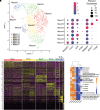
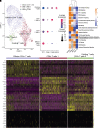
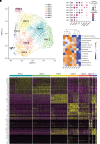
Rationale: Chronic thromboembolic pulmonary hypertension (CTEPH) is a sequela of acute pulmonary embolism (PE) in which the PE remodels into a chronic scar in the pulmonary arteries. This results in vascular obstruction, pulmonary microvasculopathy, and pulmonary hypertension. Objectives: Our current understanding of CTEPH pathobiology is primarily derived from cell-based studies limited by the use of specific cell markers or phenotypic modulation in cell culture. Therefore, our main objective was to identify the multiple cell types that constitute CTEPH thrombusy and to study their dysfunction. Methods: Here we used single-cell RNA sequencing of tissue removed at the time of pulmonary endarterectomy surgery from five patients to identify the multiple cell types. Using in vitro assays, we analyzed differences in phenotype between CTEPH thrombus and healthy pulmonary vascular cells. We studied potential therapeutic targets in cells isolated from CTEPH thrombus. Measurements and Main Results: Single-cell RNA sequencing identified multiple cell types, including macrophages, T cells, and smooth muscle cells (SMCs), that constitute CTEPH thrombus. Notably, multiple macrophage subclusters were identified but broadly split into two categories, with the larger group characterized by an upregulation of inflammatory signaling predicted to promote pulmonary vascular remodeling. CD4+ and CD8+ T cells were identified and likely contribute to chronic inflammation in CTEPH. SMCs were a heterogeneous population, with a cluster of myofibroblasts that express markers of fibrosis and are predicted to arise from other SMC clusters based on pseudotime analysis. Additionally, cultured endothelial, smooth muscle, and myofibroblast cells isolated from CTEPH fibrothrombotic material have distinct phenotypes from control cells with regard to angiogenic potential and rates of proliferation and apoptosis. Last, our analysis identified PAR1 (protease-activated receptor 1) as a potential therapeutic target that links thrombosis to chronic PE in CTEPH, with PAR1 inhibition decreasing SMC and myofibroblast proliferation and migration. Conclusions: These findings suggest a model for CTEPH similar to atherosclerosis, with chronic inflammation promoted by macrophages and T cells driving vascular remodeling through SMC modulation, and suggest new approaches for pharmacologically targeting this disease.
Keywords: CTEPH; macrophages; protease-activated receptor 1; single-cell RNA sequencing; smooth muscle cells.
Figures



Comment in
-
The Cellular Landscape of Chronic Thromboembolic Pulmonary Hypertension Revealed by Single-Cell Sequencing: Therapeutic Implications?Am J Respir Crit Care Med. 2023 May 15;207(10):1266-1268. doi: 10.1164/rccm.202302-0326ED. Am J Respir Crit Care Med. 2023. PMID: 36952236 Free PMC article. No abstract available.
References
-
- Lang IM, Dorfmüller P, Vonk Noordegraaf A. The pathobiology of chronic thromboembolic pulmonary hypertension. Ann Am Thorac Soc . 2016;13:S215–S221. - PubMed
-
- Delcroix M, Torbicki A, Gopalan D, Sitbon O, Klok FA, Lang I, et al. ERS statement on chronic thromboembolic pulmonary hypertension. Eur Respir J . 2021;57:2002828. - PubMed
-
- Wagenvoort CA. Pathology of pulmonary thromboembolism. Chest . 1995;107:10S–17S. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

