UHMK1 is a novel splicing regulatory kinase
- PMID: 36803961
- PMCID: PMC10033318
- DOI: 10.1016/j.jbc.2023.103041
UHMK1 is a novel splicing regulatory kinase
Abstract
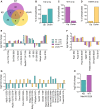
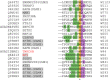
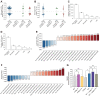
The U2AF Homology Motif Kinase 1 (UHMK1) is the only kinase that contains the U2AF homology motif, a common protein interaction domain among splicing factors. Through this motif, UHMK1 interacts with the splicing factors SF1 and SF3B1, known to participate in the 3' splice site recognition during the early steps of spliceosome assembly. Although UHMK1 phosphorylates these splicing factors in vitro, the involvement of UHMK1 in RNA processing has not previously been demonstrated. Here, we identify novel putative substrates of this kinase and evaluate UHMK1 contribution to overall gene expression and splicing, by integrating global phosphoproteomics, RNA-seq, and bioinformatics approaches. Upon UHMK1 modulation, 163 unique phosphosites were differentially phosphorylated in 117 proteins, of which 106 are novel potential substrates of this kinase. Gene Ontology analysis showed enrichment of terms previously associated with UHMK1 function, such as mRNA splicing, cell cycle, cell division, and microtubule organization. The majority of the annotated RNA-related proteins are components of the spliceosome but are also involved in several steps of gene expression. Comprehensive analysis of splicing showed that UHMK1 affected over 270 alternative splicing events. Moreover, splicing reporter assay further supported UHMK1 function on splicing. Overall, RNA-seq data demonstrated that UHMK1 knockdown had a minor impact on transcript expression and pointed to UHMK1 function in epithelial-mesenchymal transition. Functional assays demonstrated that UHMK1 modulation affects proliferation, colony formation, and migration. Taken together, our data implicate UHMK1 as a splicing regulatory kinase, connecting protein regulation through phosphorylation and gene expression in key cellular processes.
Keywords: KIS; RNA processing; RNA splicing; RNA-seq; SF1; SUGP1; UHM motif; gene expression; phosphoproteomics; protein kinase.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures






References
-
- Maucuer A., Le Caer J.P., Manceau V., Sobel A. Specific Ser-Pro phosphorylation by the RNA-recognition motif containing kinase KIS. Eur. J. Biochem. 2000;267:4456–4464. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

