Proteome-wide Mendelian randomization implicates nephronectin as an actionable mediator of the effect of obesity on COVID-19 severity
- PMID: 36805566
- PMCID: PMC9940690
- DOI: 10.1038/s42255-023-00742-w
Proteome-wide Mendelian randomization implicates nephronectin as an actionable mediator of the effect of obesity on COVID-19 severity
Abstract
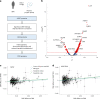
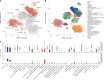
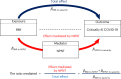
Obesity is a major risk factor for Coronavirus disease (COVID-19) severity; however, the mechanisms underlying this relationship are not fully understood. As obesity influences the plasma proteome, we sought to identify circulating proteins mediating the effects of obesity on COVID-19 severity in humans. Here, we screened 4,907 plasma proteins to identify proteins influenced by body mass index using Mendelian randomization. This yielded 1,216 proteins, whose effect on COVID-19 severity was assessed, again using Mendelian randomization. We found that an s.d. increase in nephronectin (NPNT) was associated with increased odds of critically ill COVID-19 (OR = 1.71, P = 1.63 × 10-10). The effect was driven by an NPNT splice isoform. Mediation analyses supported NPNT as a mediator. In single-cell RNA-sequencing, NPNT was expressed in alveolar cells and fibroblasts of the lung in individuals who died of COVID-19. Finally, decreasing body fat mass and increasing fat-free mass were found to lower NPNT levels. These findings provide actionable insights into how obesity influences COVID-19 severity.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
J.B.R. has served as an advisor to GlaxoSmithKline and Deerfield Capital. The institution of J.B.R. has received investigator-initiated grant funding from Eli Lilly, GlaxoSmithKline and Biogen for projects unrelated to this research. J.B.R. is the CEO of 5 Prime Sciences (
Figures





Comment in
-
Actionable targets to reduce COVID-19 severity.Nat Metab. 2023 Feb;5(2):195-196. doi: 10.1038/s42255-023-00743-9. Nat Metab. 2023. PMID: 36805565 No abstract available.
References
-
- Johns Hopkins University. COVID-19 Global Maphttps://coronavirus.jhu.edu/map.html (2022).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

