Methylation of nonessential genes in cutaneous melanoma - Rule Out hypothesis
- PMID: 36805567
- PMCID: PMC10148896
- DOI: 10.1097/CMR.0000000000000881
Methylation of nonessential genes in cutaneous melanoma - Rule Out hypothesis
Abstract
Differential methylation plays an important role in melanoma development and is associated with survival, progression and response to treatment. However, the mechanisms by which methylation promotes melanoma development are poorly understood. The traditional explanation of selective advantage provided by differential methylation postulates that hypermethylation of regulatory 5'-cytosine-phosphate-guanine-3' dinucleotides (CpGs) downregulates the expression of tumor suppressor genes and therefore promotes tumorigenesis. We believe that other (not necessarily alternative) explanations of the selective advantages of methylation are also possible. Here, we hypothesize that melanoma cells use methylation to shut down transcription of nonessential genes - those not required for cell survival and proliferation. Suppression of nonessential genes allows tumor cells to be more efficient in terms of energy and resource usage, providing them with a selective advantage over the tumor cells that transcribe and subsequently translate genes they do not need. We named the hypothesis the Rule Out (RO) hypothesis. The RO hypothesis predicts higher methylation of CpGs located in regulatory regions (CpG islands) of nonessential genes. It also predicts the higher methylation of regulatory CpGs linked to nonessential genes in melanomas compared to nevi and lower expression of nonessential genes in malignant (derived from melanoma) versus normal (derived from nonaffected skin) melanocytes. The analyses conducted using in-house and publicly available data found that all predictions derived from the RO hypothesis hold, providing observational support for the hypothesis.
Copyright © 2023 Wolters Kluwer Health, Inc. All rights reserved.
Conflict of interest statement
Conflict of interest
No potential conflicts of interest were disclosed.
Figures
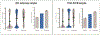



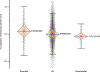

References
-
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet 2006; 7 (1):21–33. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

