Structure and function of microbial α-l-fucosidases: a mini review
- PMID: 36805644
- PMCID: PMC10154630
- DOI: 10.1042/EBC20220158
Structure and function of microbial α-l-fucosidases: a mini review
Abstract
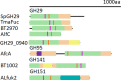
Fucose is a monosaccharide commonly found in mammalian, insect, microbial and plant glycans. The removal of terminal α-l-fucosyl residues from oligosaccharides and glycoconjugates is catalysed by α-l-fucosidases. To date, glycoside hydrolases (GHs) with exo-fucosidase activity on α-l-fucosylated substrates (EC 3.2.1.51, EC 3.2.1.-) have been reported in the GH29, GH95, GH139, GH141 and GH151 families of the Carbohydrate Active Enzymes (CAZy) database. Microbes generally encode several fucosidases in their genomes, often from more than one GH family, reflecting the high diversity of naturally occuring fucosylated structures they encounter. Functionally characterised microbial α-l-fucosidases have been shown to act on a range of substrates with α-1,2, α-1,3, α-1,4 or α-1,6 fucosylated linkages depending on the GH family and microorganism. Fucosidases show a modular organisation with catalytic domains of GH29 and GH151 displaying a (β/α)8-barrel fold while GH95 and GH141 show a (α/α)6 barrel and parallel β-helix fold, respectively. A number of crystal structures have been solved in complex with ligands, providing structural basis for their substrate specificity. Fucosidases can also be used in transglycosylation reactions to synthesise oligosaccharides. This mini review provides an overview of the enzymatic and structural properties of microbial α-l-fucosidases and some insights into their biological function and biotechnological applications.
Keywords: carbohydrate-active enzymes; fucose; fucosidases; glycoside hydrolases; gut bacteria.
© 2023 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- BB/M029042/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/F/00044452/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/F/000PR10353/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/R012490/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

