FUS regulates a subset of snoRNA expression and modulates the level of rRNA modifications
- PMID: 36806717
- PMCID: PMC9941101
- DOI: 10.1038/s41598-023-30068-2
FUS regulates a subset of snoRNA expression and modulates the level of rRNA modifications
Abstract
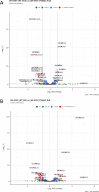
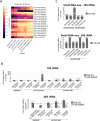
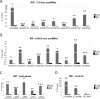
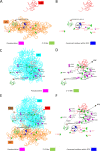
FUS is a multifunctional protein involved in many aspects of RNA metabolism, including transcription, splicing, translation, miRNA processing, and replication-dependent histone gene expression. In this work, we show that FUS depletion results in the differential expression of numerous small nucleolar RNAs (snoRNAs) that guide 2'-O methylation (2'-O-Me) and pseudouridylation of specific positions in ribosomal RNAs (rRNAs) and small nuclear RNAs (snRNAs). Using RiboMeth-seq and HydraPsiSeq for the profiling of 2'-O-Me and pseudouridylation status of rRNA species, we demonstrated considerable hypermodification at several sites in HEK293T and SH-SY5Y cells with FUS knockout (FUS KO) compared to wild-type cells. We observed a similar direction of changes in rRNA modification in differentiated SH-SY5Y cells with the FUS mutation (R495X) related to the severe disease phenotype of amyotrophic lateral sclerosis (ALS). Furthermore, the pattern of modification of some rRNA positions was correlated with the abundance of corresponding guide snoRNAs in FUS KO and FUS R495X cells. Our findings reveal a new role for FUS in modulating the modification pattern of rRNA molecules, that in turn might generate ribosome heterogeneity and constitute a fine-tuning mechanism for translation efficiency/fidelity. Therefore, we suggest that increased levels of 2'-O-Me and pseudouridylation at particular positions in rRNAs from cells with the ALS-linked FUS mutation may represent a possible new translation-related mechanism that underlies disease development and progression.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

