Evidence of structural rearrangements in ESBL-positive pESI(like) megaplasmids of S.Infantis
- PMID: 36806934
- PMCID: PMC9990980
- DOI: 10.1093/femsle/fnad014
Evidence of structural rearrangements in ESBL-positive pESI(like) megaplasmids of S.Infantis
Abstract
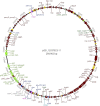
The increasing prevalence of pESI(like)-positive, multidrug-resistant (MDR) S. Infantis in Europe is a cause of major concern. As previously demonstrated, the pESI(like) megaplasmid is not only a carrier of antimicrobial resistant (AMR) genes (at least tet, dfr, and sul genes), but also harbours several virulence and fitness genes, and toxin/antitoxin systems that enhance its persistence in the S. Infantis host. In this study, five prototype pESI(like) plasmids, of either CTX-M-1 or CTX-M-65 ESBL-producing strains, were long-read sequenced using Oxford Nanopore Technology (ONT), and their complete sequences were resolved. Comparison of the structure and gene content of the five sequenced plasmids, and further comparison with previously published pESI(like) sequences, indicated that although the sequence of such pESI(like) 'mosaic' plasmids remains almost identical, their structures appear different and composed of regions inserted or transposed after different events. The results obtained in this study are essential to better understand the plasticity and the evolution of the pESI(like) megaplasmid, and therefore to better address risk management options and policy decisions to fight against AMR and MDR in Salmonella and other food-borne pathogens. Graphical representation of the pESI-like plasmid complete sequence (ID 12037823/11). Block colours indicate the function of the genes: red: repB gene; pink: class I integrons (IntI); yellow; mobile elements; blue: resistance genes; green: toxin/anti-toxin systems; grey: mer operon; light green: genes involve in conjugation.
Keywords: Salmonella Infantis; ESBL-producing Salmonella; antibiotic resistance; long read sequencing; megaplasmid; rearrangements.
© The Author(s) 2023. Published by Oxford University Press on behalf of FEMS.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

