Interleukin-23 receptor signaling impairs the stability and function of colonic regulatory T cells
- PMID: 36807140
- PMCID: PMC10432575
- DOI: 10.1016/j.celrep.2023.112128
Interleukin-23 receptor signaling impairs the stability and function of colonic regulatory T cells
Abstract
The cytokine interleukin-23 (IL-23) is involved in the pathogenesis of inflammatory and autoimmune conditions including inflammatory bowel disease (IBD). IL23R is enriched in intestinal Tregs, yet whether IL-23 modulates intestinal Tregs remains unknown. Here, investigating IL-23R signaling in Tregs specifically, we show that colonic Tregs highly express Il23r compared with Tregs from other compartments and their frequency is reduced upon IL-23 administration and impairs Treg suppressive function. Similarly, colonic Treg frequency is increased in mice lacking Il23r specifically in Tregs and exhibits a competitive advantage over IL-23R-sufficient Tregs during inflammation. Finally, IL-23 antagonizes liver X receptor pathway, cellular cholesterol transporter Abca1, and increases Treg apoptosis. Our results show that IL-23R signaling regulates intestinal Tregs by increasing cell turnover, antagonizing suppression, and decreasing cholesterol efflux. These results suggest that IL-23 negatively regulates Tregs in the intestine with potential implications for promoting chronic inflammation in patients with IBD.
Keywords: CP: Immunology; IBD; IL-23R12; Treg cell; Tregs; colitis; colon; interleukin-23; ustekinumab.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
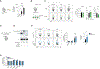
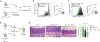

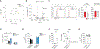
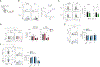
References
-
- Beaudoin M, Goyette P, Boucher G, Lo KS, Rivas MA, Stevens C, Alikashani A, Ladouceur M, Ellinghaus D, Törkvist L, et al. (2013). Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet. 9, e1003723. 10.1371/journal.pgen.1003723. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

