Molecular Mechanisms of MmpL3 Function and Inhibition
- PMID: 36809064
- PMCID: PMC10171966
- DOI: 10.1089/mdr.2021.0424
Molecular Mechanisms of MmpL3 Function and Inhibition
Abstract
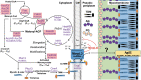
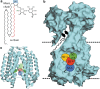
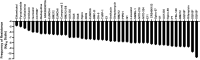
Mycobacteria species include a large number of pathogenic organisms such as Mycobacterium tuberculosis, Mycobacterium leprae, and various non-tuberculous mycobacteria. Mycobacterial membrane protein large 3 (MmpL3) is an essential mycolic acid and lipid transporter required for growth and cell viability. In the last decade, numerous studies have characterized MmpL3 with respect to protein function, localization, regulation, and substrate/inhibitor interactions. This review summarizes new findings in the field and seeks to assess future areas of research in our rapidly expanding understanding of MmpL3 as a drug target. An atlas of known MmpL3 mutations that provide resistance to inhibitors is presented, which maps amino acid substitutions to specific structural domains of MmpL3. In addition, chemical features of distinct classes of Mmpl3 inhibitors are compared to provide insights into shared and unique features of varied MmpL3 inhibitors.
Keywords: MmpL3; Mycobacterium tuberculosis; phenotypic drug discovery.
Conflict of interest statement
R.B.A. is the founder and owner of Tarn Biosciences, Inc., a company that is working to develop new antimycobacterial drugs.
Figures


References
-
- WHO. Global Tuberculosis Report. WHO: Genava; 2021.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
