Live Cell Imaging Reveals HBV Capsid Translocation from the Nucleus To the Cytoplasm Enabled by Cell Division
- PMID: 36809075
- PMCID: PMC10127671
- DOI: 10.1128/mbio.03303-22
Live Cell Imaging Reveals HBV Capsid Translocation from the Nucleus To the Cytoplasm Enabled by Cell Division
Abstract
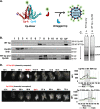
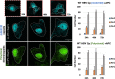
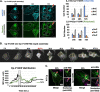
Hepatitis B virus (HBV) capsid assembly is traditionally thought to occur predominantly in the cytoplasm, where the virus gains access to the virion egress pathway. To better define sites of HBV capsid assembly, we carried out single cell imaging of HBV Core protein (Cp) subcellular trafficking over time under conditions supporting genome packaging and reverse transcription in Huh7 hepatocellular carcinoma cells. Time-course analyses including live cell imaging of fluorescently tagged Cp derivatives showed Cp to accumulate in the nucleus at early time points (~24 h), followed by a marked re-distribution to the cytoplasm at 48 to 72 h. Nucleus-associated Cp was confirmed to be capsid and/or high-order assemblages using a novel dual label immunofluorescence strategy. Nuclear-to-cytoplasmic re-localization of Cp occurred predominantly during nuclear envelope breakdown in conjunction with cell division, followed by strong cytoplasmic retention of Cp. Blocking cell division resulted in strong nuclear entrapment of high-order assemblages. A Cp mutant, Cp-V124W, predicted to exhibit enhanced assembly kinetics, also first trafficked to the nucleus to accumulate at nucleoli, consistent with the hypothesis that Cp's transit to the nucleus is a strong and constitutive process. Taken together, these results provide support for the nucleus as an early-stage site of HBV capsid assembly, and provide the first dynamic evidence of cytoplasmic retention after cell division as a mechanism underpinning capsid nucleus-to-cytoplasm relocalization. IMPORTANCE Hepatitis B virus (HBV) is an enveloped, reverse-transcribing DNA virus that is a major cause of liver disease and hepatocellular carcinoma. Subcellular trafficking events underpinning HBV capsid assembly and virion egress remain poorly characterized. Here, we developed a combination of fixed and long-term (>24 h) live cell imaging technologies to study the single cell trafficking dynamics of the HBV Core Protein (Cp). We demonstrate that Cp first accumulates in the nucleus, and forms high-order structures consistent with capsids, with the predominant route of nuclear egress being relocalization to the cytoplasm during cell division in conjunction with nuclear membrane breakdown. Single cell video microscopy demonstrated unequivocally that Cp's localization to the nucleus is constitutive. This study represents a pioneering application of live cell imaging to study HBV subcellular transport, and demonstrates links between HBV Cp and the cell cycle.
Keywords: core protein; hepatitis B virus; live cell imaging; nuclear export; subcellular trafficking; virus assembly.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

