Synergy between Genome Mining, Metabolomics, and Bioinformatics Uncovers Antibacterial Chlorinated Carbazole Alkaloids and Their Biosynthetic Gene Cluster from Streptomyces tubbatahanensis sp. nov., a Novel Actinomycete Isolated from Sulu Sea, Philippines
- PMID: 36809153
- PMCID: PMC10100901
- DOI: 10.1128/spectrum.03661-22
Synergy between Genome Mining, Metabolomics, and Bioinformatics Uncovers Antibacterial Chlorinated Carbazole Alkaloids and Their Biosynthetic Gene Cluster from Streptomyces tubbatahanensis sp. nov., a Novel Actinomycete Isolated from Sulu Sea, Philippines
Abstract
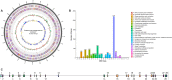
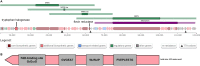
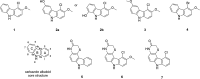
In this study, a novel actinomycete strain, DSD3025T, isolated from the underexplored marine sediments in Tubbataha Reefs Natural Park, Sulu Sea, Philippines, with the proposed name Streptomyces tubbatahanensis sp. nov., was described using polyphasic approaches and characterized using whole-genome sequencing. Its specialized metabolites were profiled using mass spectrometry and nuclear magnetic resonance analyses, followed by antibacterial, anticancer, and toxicity screening. The S. tubbatahanensis DSD3025T genome was comprised of 7.76 Mbp with a 72.3% G+C content. The average nucleotide identity and digital DNA-DNA hybridization values were 96.5% and 64.1%, respectively, compared with its closest related species, thus delineating the novelty of Streptomyces species. The genome encoded 29 putative biosynthetic gene clusters (BGCs), including a BGC region containing tryptophan halogenase and its associated flavin reductase, which were not found in its close Streptomyces relatives. The metabolite profiling unfolded six rare halogenated carbazole alkaloids, with chlocarbazomycin A as the major compound. A biosynthetic pathway for chlocarbazomycin A was proposed using genome mining, metabolomics, and bioinformatics platforms. Chlocarbazomycin A produced by S. tubbatahanensis DSD3025T has antibacterial activities against Staphylococcus aureus ATCC BAA-44 and Streptococcus pyogenes and showed antiproliferative activity against colon (HCT-116) and ovarian (A2780) human cancer cell lines. Chlocarbazomycin A exhibited no toxicity to liver cells but moderate and high toxicity to kidney and cardiac cell lines, respectively. IMPORTANCE Streptomyces tubbatahanensis DSD3025T is a novel actinomycete with antibiotic and anticancer activities from Tubbataha Reefs Natural Park, a United Nations Educational, Scientific and Cultural Organization World Heritage Site in Sulu Sea and considered one of the Philippines' oldest and most-well-protected marine ecosystems. In silico genome mining tools were used to identify putative BGCs that led to the discovery of genes involved in the production of halogenated carbazole alkaloids and new natural products. By integrating bioinformatics-driven genome mining and metabolomics, we unearthed the hidden biosynthetic richness and mined the associated chemical entities from the novel Streptomyces species. The bioprospecting of novel Streptomyces species from marine sediments of underexplored ecological niches serves as an important source of antibiotic and anticancer drug leads with unique chemical scaffolds.
Keywords: Streptomyces; antibiotic; anticancer; biosynthetic gene clusters; flavin reductase; halogenated carbazole alkaloids; specialized metabolites; tryptophan halogenase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- World Health Organization. 2021. Antimicrobial resistance. https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance.
-
- Lo Grasso L, Chilura-Martino D, Alduina R. 2016. Production of antibacterial compounds from Actinomycetes, p 272–282. In Dhanasekaran D, Jiang Y (ed), Actinobacteria: basics and biotechological applications. IntechOpen. doi: 10.5772/60457. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

