Merlin tumor suppressor function is regulated by PIP2-mediated dimerization
- PMID: 36809290
- PMCID: PMC9942953
- DOI: 10.1371/journal.pone.0281876
Merlin tumor suppressor function is regulated by PIP2-mediated dimerization
Abstract
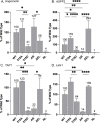
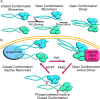
Neurofibromatosis Type 2 is an inherited disease characterized by Schwann cell tumors of cranial and peripheral nerves. The NF2 gene encodes Merlin, a member of the ERM family consisting of an N-terminal FERM domain, a central α-helical region, and a C-terminal domain. Changes in the intermolecular FERM-CTD interaction allow Merlin to transition between an open, FERM accessible conformation and a closed, FERM-inaccessible conformation, modulating Merlin activity. Merlin has been shown to dimerize, but the regulation and function Merlin dimerization is not clear. We used a nanobody based binding assay to show that Merlin dimerizes via a FERM-FERM interaction, orientated with each C-terminus close to each other. Patient derived and structural mutants show that dimerization controls interactions with specific binding partners, including HIPPO pathway components, and correlates with tumor suppressor activity. Gel filtration experiments showed that dimerization occurs after a PIP2 mediated transition from closed to open conformation monomers. This process requires the first 18 amino acids of the FERM domain and is inhibited by phosphorylation at serine 518. The discovery that active, open conformation Merlin is a dimer represents a new paradigm for Merlin function with implications for the development of therapies designed to compensate for Merlin loss.
Copyright: © 2023 Hennigan et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Evans DG, Huson SM, Donnai D, Neary W, Blair V, Newton V, et al. A clinical study of type 2 neurofibromatosis. The Quarterly journal of medicine. 1992;84(304):603–18. . - PubMed
-
- Gutmann DH, Aylsworth A, Carey JC, Korf B, Marks J, Pyeritz RE, et al. The diagnostic evaluation and multidisciplinary management of neurofibromatosis 1 and neurofibromatosis 2. JAMA: the journal of the American Medical Association. 1997;278(1):51–7. . - PubMed
-
- Morrison H, Sherman LS, Legg J, Banine F, Isacke C, Haipek CA, et al. The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes & development. 2001;15(8):968–80. doi: 10.1101/gad.189601 ; PubMed Central PMCID: PMC312675. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

