Off-target piRNA gene silencing in Drosophila melanogaster rescued by a transposable element insertion
- PMID: 36809339
- PMCID: PMC9983838
- DOI: 10.1371/journal.pgen.1010598
Off-target piRNA gene silencing in Drosophila melanogaster rescued by a transposable element insertion
Abstract
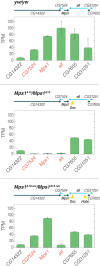
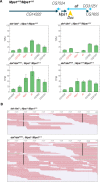
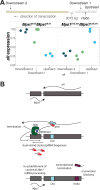
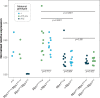
Transposable elements (TE) are selfish genetic elements that can cause harmful mutations. In Drosophila, it has been estimated that half of all spontaneous visible marker phenotypes are mutations caused by TE insertions. Several factors likely limit the accumulation of exponentially amplifying TEs within genomes. First, synergistic interactions between TEs that amplify their harm with increasing copy number are proposed to limit TE copy number. However, the nature of this synergy is poorly understood. Second, because of the harm posed by TEs, eukaryotes have evolved systems of small RNA-based genome defense to limit transposition. However, as in all immune systems, there is a cost of autoimmunity and small RNA-based systems that silence TEs can inadvertently silence genes flanking TE insertions. In a screen for essential meiotic genes in Drosophila melanogaster, a truncated Doc retrotransposon within a neighboring gene was found to trigger the germline silencing of ald, the Drosophila Mps1 homolog, a gene essential for proper chromosome segregation in meiosis. A subsequent screen for suppressors of this silencing identified a new insertion of a Hobo DNA transposon in the same neighboring gene. Here we describe how the original Doc insertion triggers flanking piRNA biogenesis and local gene silencing. We show that this local gene silencing occurs in cis and is dependent on deadlock, a component of the Rhino-Deadlock-Cutoff (RDC) complex, to trigger dual-strand piRNA biogenesis at TE insertions. We further show how the additional Hobo insertion leads to de-silencing by reducing flanking piRNA biogenesis triggered by the original Doc insertion. These results support a model of TE-mediated gene silencing by piRNA biogenesis in cis that depends on local determinants of transcription. This may explain complex patterns of off-target gene silencing triggered by TEs within populations and in the laboratory. It also provides a mechanism of sign epistasis among TE insertions, illuminates the complex nature of their interactions and supports a model in which off-target gene silencing shapes the evolution of the RDC complex.
Copyright: © 2023 Miller et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Green MM. Mobile DNA elements and spontaneous gene mutation. Banbury Reports. Cold Spring Harbor Laboratory; 1988. pp. 41–50.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

