Organ and cell-specific biomarkers of Long-COVID identified with targeted proteomics and machine learning
- PMID: 36809921
- PMCID: PMC9942653
- DOI: 10.1186/s10020-023-00610-z
Organ and cell-specific biomarkers of Long-COVID identified with targeted proteomics and machine learning
Abstract
Background: Survivors of acute COVID-19 often suffer prolonged, diffuse symptoms post-infection, referred to as "Long-COVID". A lack of Long-COVID biomarkers and pathophysiological mechanisms limits effective diagnosis, treatment and disease surveillance. We performed targeted proteomics and machine learning analyses to identify novel blood biomarkers of Long-COVID.
Methods: A case-control study comparing the expression of 2925 unique blood proteins in Long-COVID outpatients versus COVID-19 inpatients and healthy control subjects. Targeted proteomics was accomplished with proximity extension assays, and machine learning was used to identify the most important proteins for identifying Long-COVID patients. Organ system and cell type expression patterns were identified with Natural Language Processing (NLP) of the UniProt Knowledgebase.
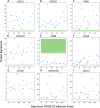
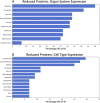
Results: Machine learning analysis identified 119 relevant proteins for differentiating Long-COVID outpatients (Bonferonni corrected P < 0.01). Protein combinations were narrowed down to two optimal models, with nine and five proteins each, and with both having excellent sensitivity and specificity for Long-COVID status (AUC = 1.00, F1 = 1.00). NLP expression analysis highlighted the diffuse organ system involvement in Long-COVID, as well as the involved cell types, including leukocytes and platelets, as key components associated with Long-COVID.
Conclusions: Proteomic analysis of plasma from Long-COVID patients identified 119 highly relevant proteins and two optimal models with nine and five proteins, respectively. The identified proteins reflected widespread organ and cell type expression. Optimal protein models, as well as individual proteins, hold the potential for accurate diagnosis of Long-COVID and targeted therapeutics.
Keywords: Biomarker; COVID-19; Cell types; Long-COVID; Machine learning; Organ system; Targeted proteomics.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

