Bovine meat and milk factor protein expression in tumor-free mucosa of colorectal cancer patients coincides with macrophages and might interfere with patient survival
- PMID: 36811271
- PMCID: PMC11076986
- DOI: 10.1002/1878-0261.13390
Bovine meat and milk factor protein expression in tumor-free mucosa of colorectal cancer patients coincides with macrophages and might interfere with patient survival
Abstract
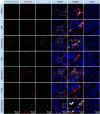
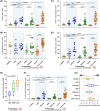
Bovine milk and meat factors (BMMFs) are plasmid-like DNA molecules isolated from bovine milk and serum, as well as the peritumor of colorectal cancer (CRC) patients. BMMFs have been proposed as zoonotic infectious agents and drivers of indirect carcinogenesis of CRC, inducing chronic tissue inflammation, radical formation and increased levels of DNA damage. Data on expression of BMMFs in large clinical cohorts to test an association with co-markers and clinical parameters were not previously available and were therefore assessed in this study. Tissue sections with paired tumor-adjacent mucosa and tumor tissues of CRC patients [individual cohorts and tissue microarrays (TMAs) (n = 246)], low-/high-grade dysplasia (LGD/HGD) and mucosa of healthy donors were used for immunohistochemical quantification of the expression of BMMF replication protein (Rep) and CD68/CD163 (macrophages) by co-immunofluorescence microscopy and immunohistochemical scoring (TMA). Rep was expressed in the tumor-adjacent mucosa of 99% of CRC patients (TMA), was histologically associated with CD68+/CD163+ macrophages and was increased in CRC patients when compared to healthy controls. Tumor tissues showed only low stromal Rep expression. Rep was expressed in LGD and less in HGD but was strongly expressed in LGD/HGD-adjacent tissues. Albeit not reaching statistical significance, incidence curves for CRC-specific death were increased for higher Rep expression (TMA), with high tumor-adjacent Rep expression being linked to the highest incidence of death. BMMF Rep expression might represent a marker and early risk factor for CRC. The correlation between Rep and CD68 expression supports a previous hypothesis that BMMF-specific inflammatory regulations, including macrophages, are involved in the pathogenesis of CRC.
Keywords: bovine meat and milk factors; chronic inflammation; colorectal cancer; infectious indirect carcinogenesis.
© 2023 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Current Research on Small Circular Molecules: A Comprehensive Overview on SPHINX/BMMF.Genes (Basel). 2024 May 24;15(6):678. doi: 10.3390/genes15060678. Genes (Basel). 2024. PMID: 38927614 Free PMC article. Review.
-
Bovine Meat and Milk Factor (BMMF) Protein Is Expressed in Macrophages Spread Widely over the Mucosa of Colorectal Cancer Patients.Cells. 2025 Mar 19;14(6):455. doi: 10.3390/cells14060455. Cells. 2025. PMID: 40136704 Free PMC article.
-
Analysis of chronic inflammatory lesions of the colon for BMMF Rep antigen expression and CD68 macrophage interactions.Proc Natl Acad Sci U S A. 2021 Mar 23;118(12):e2025830118. doi: 10.1073/pnas.2025830118. Proc Natl Acad Sci U S A. 2021. PMID: 33723077 Free PMC article.
-
Expression of Bovine Meat and Milk Factor in Hepatocellular Carcinoma and Colorectal Liver Metastasis Patients.J Med Virol. 2025 Aug;97(8):e70507. doi: 10.1002/jmv.70507. J Med Virol. 2025. PMID: 40741925
-
Structural expression of bovine milk and meat factors in tissues of colorectal, lung and pancreatic cancer patients.Int J Cancer. 2023 Jul 1;153(1):173-182. doi: 10.1002/ijc.34374. Epub 2022 Dec 8. Int J Cancer. 2023. PMID: 36444499
Cited by
-
Current Research on Small Circular Molecules: A Comprehensive Overview on SPHINX/BMMF.Genes (Basel). 2024 May 24;15(6):678. doi: 10.3390/genes15060678. Genes (Basel). 2024. PMID: 38927614 Free PMC article. Review.
-
First Insights into the Occurrence of Circular Single-Stranded DNA Genomes in Asian and African Cattle.Animals (Basel). 2023 Apr 27;13(9):1492. doi: 10.3390/ani13091492. Animals (Basel). 2023. PMID: 37174530 Free PMC article.
-
Bovine Meat and Milk Factor (BMMF) Protein Is Expressed in Macrophages Spread Widely over the Mucosa of Colorectal Cancer Patients.Cells. 2025 Mar 19;14(6):455. doi: 10.3390/cells14060455. Cells. 2025. PMID: 40136704 Free PMC article.
-
Outer membrane vesicles of Acinetobacter baumannii DS002 carry circular DNA similar to bovine meat and milk factors (BMMFs) and SPHINX 2.36 and probably play a role in interdomain lateral gene transfer.Microbiol Spectr. 2024 Sep 3;12(9):e0081724. doi: 10.1128/spectrum.00817-24. Epub 2024 Aug 5. Microbiol Spectr. 2024. PMID: 39101807 Free PMC article.
-
Bovine Meat and Milk Factor-like Sequences Are Frequently Detected in Renal Cell Carcinoma Tissues.Cancers (Basel). 2024 Apr 29;16(9):1746. doi: 10.3390/cancers16091746. Cancers (Basel). 2024. PMID: 38730698 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

