ORIGIN OF RECOGNITION COMPLEX 3 controls the development of maternal excess endosperm in the Paspalum simplex agamic complex (Poaceae)
- PMID: 36812152
- PMCID: PMC10199125
- DOI: 10.1093/jxb/erad069
ORIGIN OF RECOGNITION COMPLEX 3 controls the development of maternal excess endosperm in the Paspalum simplex agamic complex (Poaceae)
Abstract
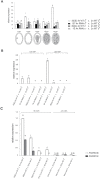
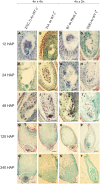
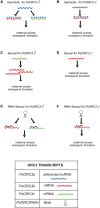
Pseudogamous apomixis in Paspalum simplex generates seeds with embryos genetically identical to the mother plant and endosperms deviating from the canonical 2(maternal):1(paternal) parental genome contribution into a maternal excess 4m:1p genome ratio. In P. simplex, the gene homologous to that coding for subunit 3 of the ORIGIN OF RECOGNITION COMPLEX (PsORC3) exists in three isogenic forms: PsORC3a is apomixis specific and constitutively expressed in developing endosperm whereas PsORCb and PsORCc are up-regulated in sexual endosperms and silenced in apomictic ones. This raises the question of how the different arrangement and expression profiles of these three ORC3 isogenes are linked to seed development in interploidy crosses generating maternal excess endosperms. We demonstrate that down-regulation of PsORC3b in sexual tetraploid plants is sufficient to restore seed fertility in interploidy 4n×2n crosses and, in turn, its expression level at the transition from proliferating to endoreduplication endosperm developmental stages dictates the fate of these seeds. Furthermore, we show that only when being maternally inherited can PsORC3c up-regulate PsORC3b. Our findings lay the basis for an innovative route-based on ORC3 manipulation-to introgress the apomictic trait into sexual crops and overcome the fertilization barriers in interploidy crosses.
Keywords: ORC3; Paspalum simplex; Apomixis; cell cycle; interploidy crosses; maternal excess endosperm.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Conflict of interest statement
The authors declare no conflict of interest
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

