Radical Transport Facilitated by a Proton Transfer Network at the Subunit Interface of Ribonucleotide Reductase
- PMID: 36812162
- PMCID: PMC10561588
- DOI: 10.1021/jacs.2c11483
Radical Transport Facilitated by a Proton Transfer Network at the Subunit Interface of Ribonucleotide Reductase
Abstract
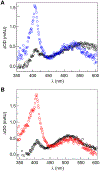
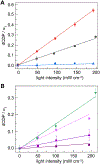
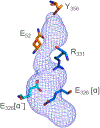
Ribonucleotide reductases (RNRs) play an essential role in the conversion of nucleotides to deoxynucleotides in all organisms. The Escherichia coli class Ia RNR requires two homodimeric subunits, α and β. The active form is an asymmetric αα'ββ' complex. The α subunit houses the site for nucleotide reduction initiated by a thiyl radical (C439•), and the β subunit houses the diferric-tyrosyl radical (Y122•) that is essential for C439• formation. The reactions require a highly regulated and reversible long-range proton-coupled electron transfer pathway involving Y122•[β] ↔ W48?[β] ↔ Y356[β] ↔ Y731[α] ↔ Y730[α] ↔ C439[α]. In a recent cryo-EM structure, Y356[β] was revealed for the first time and it, along with Y731[α], spans the asymmetric α/β interface. An E52[β] residue, which is essential for Y356 oxidation, allows access to the interface and resides at the head of a polar region comprising R331[α], E326[α], and E326[α'] residues. Mutagenesis studies with canonical and unnatural amino acid substitutions now suggest that these ionizable residues are important in enzyme activity. To gain further insights into the roles of these residues, Y356• was photochemically generated using a photosensitizer covalently attached adjacent to Y356[β]. Mutagenesis studies, transient absorption spectroscopy, and photochemical assays monitoring deoxynucleotide formation collectively indicate that the E52[β], R331[α], E326[α], and E326[α'] network plays the essential role of shuttling protons associated with Y356 oxidation from the interface to bulk solvent.
Figures



Similar articles
-
Reversible, long-range radical transfer in E. coli class Ia ribonucleotide reductase.Acc Chem Res. 2013 Nov 19;46(11):2524-35. doi: 10.1021/ar4000407. Epub 2013 Jun 4. Acc Chem Res. 2013. PMID: 23730940 Free PMC article.
-
A >200 meV Uphill Thermodynamic Landscape for Radical Transport in Escherichia coli Ribonucleotide Reductase Determined Using Fluorotyrosine-Substituted Enzymes.J Am Chem Soc. 2016 Oct 19;138(41):13706-13716. doi: 10.1021/jacs.6b08200. Epub 2016 Oct 7. J Am Chem Soc. 2016. PMID: 28068088 Free PMC article.
-
Gated Proton Release during Radical Transfer at the Subunit Interface of Ribonucleotide Reductase.J Am Chem Soc. 2021 Jan 13;143(1):176-183. doi: 10.1021/jacs.0c07879. Epub 2020 Dec 23. J Am Chem Soc. 2021. PMID: 33353307 Free PMC article.
-
Long-range proton-coupled electron transfer in the Escherichia coli class Ia ribonucleotide reductase.Essays Biochem. 2017 May 9;61(2):281-292. doi: 10.1042/EBC20160072. Print 2017 May 9. Essays Biochem. 2017. PMID: 28487404 Review.
-
The periodic table of ribonucleotide reductases.J Biol Chem. 2021 Oct;297(4):101137. doi: 10.1016/j.jbc.2021.101137. Epub 2021 Aug 27. J Biol Chem. 2021. PMID: 34461093 Free PMC article. Review.
Cited by
-
Structural Elucidation of the Reduced Mn(III)/Fe(III) Intermediate of the Radical-Initiating Metallocofactor in Chlamydia trachomatis Ribonucleotide Reductase.Biochemistry. 2025 Mar 4;64(5):1157-1167. doi: 10.1021/acs.biochem.4c00692. Epub 2025 Feb 17. Biochemistry. 2025. PMID: 39960811 Free PMC article.
-
Protein engineering a PhotoRNR chimera based on a unifying evolutionary apparatus among the natural classes of ribonucleotide reductases.Proc Natl Acad Sci U S A. 2024 Apr 30;121(18):e2317291121. doi: 10.1073/pnas.2317291121. Epub 2024 Apr 22. Proc Natl Acad Sci U S A. 2024. PMID: 38648489 Free PMC article.
-
Conformational control over proton-coupled electron transfer in metalloenzymes.Nat Rev Chem. 2024 Oct;8(10):762-775. doi: 10.1038/s41570-024-00646-7. Epub 2024 Sep 2. Nat Rev Chem. 2024. PMID: 39223400 Free PMC article. Review.
-
Proton-coupled electron transfer dynamics in the alternative oxidase.Chem Sci. 2024 Oct 11;15(44):18572-80. doi: 10.1039/d4sc05060f. Online ahead of print. Chem Sci. 2024. PMID: 39444558 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

