A nucleolar long "non-coding" RNA encodes a novel protein that functions in response to stress
- PMID: 36812203
- PMCID: PMC9992852
- DOI: 10.1073/pnas.2221109120
A nucleolar long "non-coding" RNA encodes a novel protein that functions in response to stress
Abstract
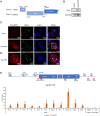
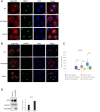
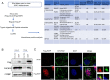
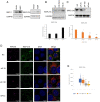
Certain long non-coding RNAs (lncRNAs) are known to contain small open reading frames that can be translated. Here we describe a much larger 25 kDa human protein, "
Keywords: lncRNA; mitochondria; nucleolus; stress.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
-
- Matsumoto A., et al. , mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 541, 228–232 (2017). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

