A proteomic survival predictor for COVID-19 patients in intensive care
- PMID: 36812516
- PMCID: PMC9931303
- DOI: 10.1371/journal.pdig.0000007
A proteomic survival predictor for COVID-19 patients in intensive care
Abstract
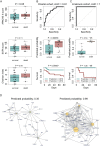
Global healthcare systems are challenged by the COVID-19 pandemic. There is a need to optimize allocation of treatment and resources in intensive care, as clinically established risk assessments such as SOFA and APACHE II scores show only limited performance for predicting the survival of severely ill COVID-19 patients. Additional tools are also needed to monitor treatment, including experimental therapies in clinical trials. Comprehensively capturing human physiology, we speculated that proteomics in combination with new data-driven analysis strategies could produce a new generation of prognostic discriminators. We studied two independent cohorts of patients with severe COVID-19 who required intensive care and invasive mechanical ventilation. SOFA score, Charlson comorbidity index, and APACHE II score showed limited performance in predicting the COVID-19 outcome. Instead, the quantification of 321 plasma protein groups at 349 timepoints in 50 critically ill patients receiving invasive mechanical ventilation revealed 14 proteins that showed trajectories different between survivors and non-survivors. A predictor trained on proteomic measurements obtained at the first time point at maximum treatment level (i.e. WHO grade 7), which was weeks before the outcome, achieved accurate classification of survivors (AUROC 0.81). We tested the established predictor on an independent validation cohort (AUROC 1.0). The majority of proteins with high relevance in the prediction model belong to the coagulation system and complement cascade. Our study demonstrates that plasma proteomics can give rise to prognostic predictors substantially outperforming current prognostic markers in intensive care.
Copyright: © 2022 Demichev et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors declare no competing interests. Author John F. Timms was unable to confirm their authorship contributions. On their behalf, the corresponding author has reported their contributions to the best of their knowledge.
Figures

References
-
- Clift AK, Coupland CAC, Keogh RH, Diaz-Ordaz K, Williamson E, Harrison EM, et al.. Living risk prediction algorithm (QCOVID) for risk of hospital admission and mortality from coronavirus 19 in adults: national derivation and validation cohort study. BMJ. 2020;371: m3731. doi: 10.1136/bmj.m3731 - DOI - PMC - PubMed
-
- Knight SR, Ho A, Pius R, Buchan I, Carson G, Drake TM, et al.. Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: development and validation of the 4C Mortality Score. BMJ. 2020;370: m3339. doi: 10.1136/bmj.m3339 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
