Automated large-scale prediction of exudative AMD progression using machine-read OCT biomarkers
- PMID: 36812608
- PMCID: PMC9931262
- DOI: 10.1371/journal.pdig.0000106
Automated large-scale prediction of exudative AMD progression using machine-read OCT biomarkers
Abstract
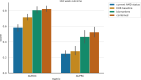
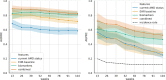
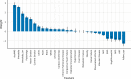
Age-related Macular Degeneration (AMD) is a major cause of irreversible vision loss in individuals over 55 years old in the United States. One of the late-stage manifestations of AMD, and a major cause of vision loss, is the development of exudative macular neovascularization (MNV). Optical Coherence Tomography (OCT) is the gold standard to identify fluid at different levels within the retina. The presence of fluid is considered the hallmark to define the presence of disease activity. Anti-vascular growth factor (anti-VEGF) injections can be used to treat exudative MNV. However, given the limitations of anti-VEGF treatment, as burdensome need for frequent visits and repeated injections to sustain efficacy, limited durability of the treatment, poor or no response, there is a great interest in detecting early biomarkers associated with a higher risk for AMD progression to exudative forms in order to optimize the design of early intervention clinical trials. The annotation of structural biomarkers on optical coherence tomography (OCT) B-scans is a laborious, complex and time-consuming process, and discrepancies between human graders can introduce variability into this assessment. To address this issue, a deep-learning model (SLIVER-net) was proposed, which could identify AMD biomarkers on structural OCT volumes with high precision and without human supervision. However, the validation was performed on a small dataset, and the true predictive power of these detected biomarkers in the context of a large cohort has not been evaluated. In this retrospective cohort study, we perform the largest-scale validation of these biomarkers to date. We also assess how these features combined with other EHR data (demographics, comorbidities, etc) affect and/or improve the prediction performance relative to known factors. Our hypothesis is that these biomarkers can be identified by a machine learning algorithm without human supervision, in a way that they preserve their predictive nature. The way we test this hypothesis is by building several machine learning models utilizing these machine-read biomarkers and assessing their added predictive power. We found that not only can we show that the machine-read OCT B-scan biomarkers are predictive of AMD progression, we also observe that our proposed combined OCT and EHR data-based algorithm outperforms the state-of-the-art solution in clinically relevant metrics and provides actionable information which has the potential to improve patient care. In addition, it provides a framework for automated large-scale processing of OCT volumes, making it possible to analyze vast archives without human supervision.
Copyright: © 2023 Rudas et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: Eran Halperin has received consulting fees from United Health Group. SriniVas Sadda has received consulting fees from Amgen, Allergan, Genentech-Roche, Oxurion, Novartis, Regeneron, Iveric, 4DMT, Centervue, Heidelberg, Optos, Carl Zeiss Meditec, Nidek, and Topcon.
Figures

Similar articles
-
Optical Coherence Tomography-Based Deep-Learning Models for Classifying Normal and Age-Related Macular Degeneration and Exudative and Non-Exudative Age-Related Macular Degeneration Changes.Ophthalmol Ther. 2019 Dec;8(4):527-539. doi: 10.1007/s40123-019-00207-y. Epub 2019 Aug 12. Ophthalmol Ther. 2019. PMID: 31407214 Free PMC article.
-
Evaluation of an Artificial Intelligence-Based Detector of Sub- and Intraretinal Fluid on a Large Set of Optical Coherence Tomography Volumes in Age-Related Macular Degeneration and Diabetic Macular Edema.Ophthalmologica. 2022;245(6):516-527. doi: 10.1159/000527345. Epub 2022 Oct 10. Ophthalmologica. 2022. PMID: 36215958
-
Automated detection of exudative age-related macular degeneration in spectral domain optical coherence tomography using deep learning.Graefes Arch Clin Exp Ophthalmol. 2018 Feb;256(2):259-265. doi: 10.1007/s00417-017-3850-3. Epub 2017 Nov 20. Graefes Arch Clin Exp Ophthalmol. 2018. PMID: 29159541
-
Lessons Learned From Avastin and OCT-The Great, the Good, the Bad, and the Ugly: The LXXV Edward Jackson Memorial Lecture.Am J Ophthalmol. 2019 Aug;204:26-45. doi: 10.1016/j.ajo.2019.02.036. Epub 2019 Mar 7. Am J Ophthalmol. 2019. PMID: 30851267 Review.
-
A paradigm shift in imaging biomarkers in neovascular age-related macular degeneration.Prog Retin Eye Res. 2016 Jan;50:1-24. doi: 10.1016/j.preteyeres.2015.07.007. Epub 2015 Aug 22. Prog Retin Eye Res. 2016. PMID: 26307399 Review.
Cited by
-
Artificial intelligence for diagnosing exudative age-related macular degeneration.Cochrane Database Syst Rev. 2024 Oct 17;10(10):CD015522. doi: 10.1002/14651858.CD015522.pub2. Cochrane Database Syst Rev. 2024. PMID: 39417312
-
Non-exudative OCT findings in neovascular AMD.Eye (Lond). 2025 Feb;39(3):516-526. doi: 10.1038/s41433-024-03461-y. Epub 2024 Nov 25. Eye (Lond). 2025. PMID: 39587331 Review.
-
Effect of tokenization on transformers for biological sequences.Bioinformatics. 2024 Mar 29;40(4):btae196. doi: 10.1093/bioinformatics/btae196. Bioinformatics. 2024. PMID: 38608190 Free PMC article.
-
Recent advances in the application of artificial intelligence in age-related macular degeneration.BMJ Open Ophthalmol. 2024 Nov 13;9(1):e001903. doi: 10.1136/bmjophth-2024-001903. BMJ Open Ophthalmol. 2024. PMID: 39537399 Free PMC article. Review.
-
Breakthroughs in diabetic retinopathy diagnosis and treatment using preclinical research models: current progress and future directions.Ann Med. 2025 Dec;57(1):2531251. doi: 10.1080/07853890.2025.2531251. Epub 2025 Jul 13. Ann Med. 2025. PMID: 40652408 Free PMC article. Review.
References
-
- Spaide RF, Jaffe GJ, Sarraf D, Freund KB, Sadda SR, Staurenghi G, et al.. Consensus Nomenclature for Reporting Neovascular Age-Related Macular Degeneration Data: Consensus on Neovascular Age-Related Macular Degeneration Nomenclature Study Group. Ophthalmology. 2020;127(5):616–36. doi: 10.1016/j.ophtha.2019.11.004 - DOI - PMC - PubMed
