An adverse tumor-protective effect of IDO1 inhibition
- PMID: 36812891
- PMCID: PMC9975322
- DOI: 10.1016/j.xcrm.2023.100941
An adverse tumor-protective effect of IDO1 inhibition
Abstract
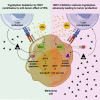
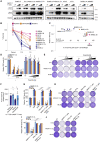
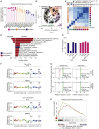
By restoring tryptophan, indoleamine 2,3-dioxygenase 1 (IDO1) inhibitors aim to reactivate anti-tumor T cells. However, a phase III trial assessing their clinical benefit failed, prompting us to revisit the role of IDO1 in tumor cells under T cell attack. We show here that IDO1 inhibition leads to an adverse protection of melanoma cells to T cell-derived interferon-gamma (IFNγ). RNA sequencing and ribosome profiling shows that IFNγ shuts down general protein translation, which is reversed by IDO1 inhibition. Impaired translation is accompanied by an amino acid deprivation-dependent stress response driving activating transcription factor-4 (ATF4)high/microphtalmia-associated transcription factor (MITF)low transcriptomic signatures, also in patient melanomas. Single-cell sequencing analysis reveals that MITF downregulation upon immune checkpoint blockade treatment predicts improved patient outcome. Conversely, MITF restoration in cultured melanoma cells causes T cell resistance. These results highlight the critical role of tryptophan and MITF in the melanoma response to T cell-derived IFNγ and uncover an unexpected negative consequence of IDO1 inhibition.
Keywords: IDO1; IDO1 inhibition; IFNgamma; MITF; T cells; clinical trial; immunotherapy; melanoma; translation.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests D.S.P. is co-founder, shareholder, and advisor of Immagene, which is unrelated to this study.
Figures


References
-
- Kotake Y., Masayama I. Studien über den intermediären Stoffwechsel des Tryptophans XVIII—XXIV. Hoppe-Seyler's. Zeitschrift für. physiologische Chemie. 1936;243:237–265. doi: 10.1515/bchm2.1936.243.6.237. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

