Bat pluripotent stem cells reveal unusual entanglement between host and viruses
- PMID: 36812912
- PMCID: PMC10085545
- DOI: 10.1016/j.cell.2023.01.011
Bat pluripotent stem cells reveal unusual entanglement between host and viruses
Abstract
Bats are distinctive among mammals due to their ability to fly, use laryngeal echolocation, and tolerate viruses. However, there are currently no reliable cellular models for studying bat biology or their response to viral infections. Here, we created induced pluripotent stem cells (iPSCs) from two species of bats: the wild greater horseshoe bat (Rhinolophus ferrumequinum) and the greater mouse-eared bat (Myotis myotis). The iPSCs from both bat species showed similar characteristics and had a gene expression profile resembling that of cells attacked by viruses. They also had a high number of endogenous viral sequences, particularly retroviruses. These results suggest that bats have evolved mechanisms to tolerate a large load of viral sequences and may have a more intertwined relationship with viruses than previously thought. Further study of bat iPSCs and their differentiated progeny will provide insights into bat biology, virus host relationships, and the molecular basis of bats' special traits.
Keywords: bats; cancer; coronaviruses; differentiation; endogenized viruses; epigenetics; evolution; host-pathogen-interactions; induced pluripotent stem cells; inflammation; virus response.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests T.P.Z., M.D., and A.G.S. are inventors on patents and patent applications on the use of bat iPS cells, owned by the Icahn School of Medicine at Mount Sinai, New York. T.P.Z. and R.A.Y. are founders and shareholders of Paratus Sciences, and R.A.Y. is a founder and shareholder of Syros Pharmaceuticals, Camp4 Therapeutics, Omega Therapeutics, and Dewpoint Therapeutics. The A.G.S. laboratory has received research support from Pfizer, Senhwa Biosciences, Kenall Manufacturing, Avimex, Johnson & Johnson, Dynavax, 7Hills Pharma, Pharmamar, ImmunityBio, Accurius, Nanocomposix, Hexamer, N-Fold LLC, Model Medicines, and Merck, outside of the reported work. A.G.S. has consulting agreements for the following companies involving cash and/or stock: Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Vaxalto, Pagoda, Accurius, Esperovax, Farmak, Applied Biological Laboratories, and Pfizer, outside of the reported work. A.G.S. is inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections and cancer, owned by the Icahn School of Medicine at Mount Sinai, New York, outside of the reported work. A.M. is the creator of Omics Bioinformatics and owns all the stocks of this company. R.P.S. has a consulting agreement with Sema4 involving cash and is also a stockholder of this company. A.P. and S.D.W.F. are employees of Microsoft Corporation. S.D.W.F. is co-founder of DIOSynVax, Ltd. and an inventor on patent applications on the design of vaccine immunogens for the prevention of virus infections, outside of the reported work.
Figures


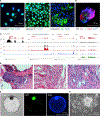


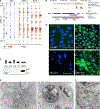
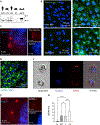
Comment in
-
Bat crazy iPSCs.Cell. 2023 Mar 2;186(5):901-903. doi: 10.1016/j.cell.2023.01.033. Cell. 2023. PMID: 36868212
-
Bat-virus entanglements.Nat Methods. 2023 Apr;20(4):483. doi: 10.1038/s41592-023-01858-3. Nat Methods. 2023. PMID: 37046017 No abstract available.
References
-
- Simmons N, and Cirranello A (2020). Bat species of the world: a taxonomic and geographic database. https://batnames.org/.
-
- Allen G (1939). Bats (Harvard University Press; ).
-
- Nagel T (1974). What is it like to be a bat. The. Philosophical Review 83, 159–168.
-
- Teeling E, Dool S, and Springer M (2012). Phylogenies, fossils and functional genes: the evolution of echolocation in bats. Evol. Hist. Bats Fossils Mol. Morphol, 1–22.
-
- Teeling EC, Vernes SC, Dávalos LM, Ray DA, Gilbert MTP, and Myers E; Bat1K Consortium (2018). Bat biology, genomes, and the Bat1K project: to generate chromosome-level genomes for all living bat species. Annu. Rev. Anim. Biosci. 6, 23–46. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

