Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase
- PMID: 36813778
- PMCID: PMC9947105
- DOI: 10.1038/s41467-023-36604-y
Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase
Abstract
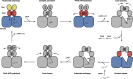
In Escherichia coli, the 14-cistron phn operon encoding carbon-phosphorus lyase allows for utilisation of phosphorus from a wide range of stable phosphonate compounds containing a C-P bond. As part of a complex, multi-step pathway, the PhnJ subunit was shown to cleave the C-P bond via a radical mechanism, however, the details of the reaction could not immediately be reconciled with the crystal structure of a 220 kDa PhnGHIJ C-P lyase core complex, leaving a significant gap in our understanding of phosphonate breakdown in bacteria. Here, we show using single-particle cryogenic electron microscopy that PhnJ mediates binding of a double dimer of the ATP-binding cassette proteins, PhnK and PhnL, to the core complex. ATP hydrolysis induces drastic structural remodelling leading to opening of the core complex and reconfiguration of a metal-binding and putative active site located at the interface between the PhnI and PhnJ subunits.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

