Mitochondrial structure and function adaptation in residual triple negative breast cancer cells surviving chemotherapy treatment
- PMID: 36813854
- PMCID: PMC10069007
- DOI: 10.1038/s41388-023-02596-8
Mitochondrial structure and function adaptation in residual triple negative breast cancer cells surviving chemotherapy treatment
Abstract
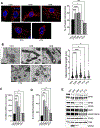
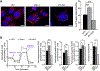
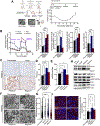
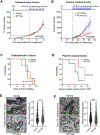
Neoadjuvant chemotherapy (NACT) used for triple negative breast cancer (TNBC) eradicates tumors in ~45% of patients. Unfortunately, TNBC patients with substantial residual cancer burden have poor metastasis free and overall survival rates. We previously demonstrated mitochondrial oxidative phosphorylation (OXPHOS) was elevated and was a unique therapeutic dependency of residual TNBC cells surviving NACT. We sought to investigate the mechanism underlying this enhanced reliance on mitochondrial metabolism. Mitochondria are morphologically plastic organelles that cycle between fission and fusion to maintain mitochondrial integrity and metabolic homeostasis. The functional impact of mitochondrial structure on metabolic output is highly context dependent. Several chemotherapy agents are conventionally used for neoadjuvant treatment of TNBC patients. Upon comparing mitochondrial effects of conventional chemotherapies, we found that DNA-damaging agents increased mitochondrial elongation, mitochondrial content, flux of glucose through the TCA cycle, and OXPHOS, whereas taxanes instead decreased mitochondrial elongation and OXPHOS. The mitochondrial effects of DNA-damaging chemotherapies were dependent on the mitochondrial inner membrane fusion protein optic atrophy 1 (OPA1). Further, we observed heightened OXPHOS, OPA1 protein levels, and mitochondrial elongation in an orthotopic patient-derived xenograft (PDX) model of residual TNBC. Pharmacologic or genetic disruption of mitochondrial fusion and fission resulted in decreased or increased OXPHOS, respectively, revealing longer mitochondria favor oxphos in TNBC cells. Using TNBC cell lines and an in vivo PDX model of residual TNBC, we found that sequential treatment with DNA-damaging chemotherapy, thus inducing mitochondrial fusion and OXPHOS, followed by MYLS22, a specific inhibitor of OPA1, was able to suppress mitochondrial fusion and OXPHOS and significantly inhibit regrowth of residual tumor cells. Our data suggest that TNBC mitochondria can optimize OXPHOS through OPA1-mediated mitochondrial fusion. These findings may provide an opportunity to overcome mitochondrial adaptations of chemoresistant TNBC.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Figures



References
-
- Liedtke C, Mazouni C, Hess KR, et al. Response to Neoadjuvant Therapy and Long-Term Survival in Patients With Triple-Negative Breast Cancer. J Clin Oncol 2008;26:1275–81. - PubMed
-
- Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet 2014;384:164–72. - PubMed
-
- von Minckwitz G, Untch M, Blohmer JU, et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol 2012;30:1796–804. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- T32 GM139534/GM/NIGMS NIH HHS/United States
- F31 CA275397/CA/NCI NIH HHS/United States
- R01 HD102149/HD/NICHD NIH HHS/United States
- K22 CA241113/CA/NCI NIH HHS/United States
- P30 DK056338/DK/NIDDK NIH HHS/United States
- R37 CA269783/CA/NCI NIH HHS/United States
- S10 OD012304/OD/NIH HHS/United States
- R01 DK114356/DK/NIDDK NIH HHS/United States
- R01 HL130249/HL/NHLBI NIH HHS/United States
- T32 GM136560/GM/NIGMS NIH HHS/United States
- P30 ES029067/ES/NIEHS NIH HHS/United States
- P30 ES030285/ES/NIEHS NIH HHS/United States
- P30 CA125123/CA/NCI NIH HHS/United States
- UM1 HG006348/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources

