Autoimmunity in Down's syndrome via cytokines, CD4 T cells and CD11c+ B cells
- PMID: 36813963
- PMCID: PMC9945839
- DOI: 10.1038/s41586-023-05736-y
Autoimmunity in Down's syndrome via cytokines, CD4 T cells and CD11c+ B cells
Abstract
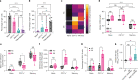
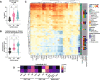
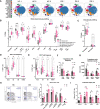
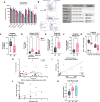
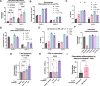
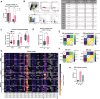
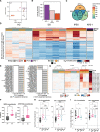
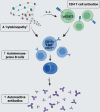
Down's syndrome (DS) presents with a constellation of cardiac, neurocognitive and growth impairments. Individuals with DS are also prone to severe infections and autoimmunity including thyroiditis, type 1 diabetes, coeliac disease and alopecia areata1,2. Here, to investigate the mechanisms underlying autoimmune susceptibility, we mapped the soluble and cellular immune landscape of individuals with DS. We found a persistent elevation of up to 22 cytokines at steady state (at levels often exceeding those in patients with acute infection) and detected basal cellular activation: chronic IL-6 signalling in CD4 T cells and a high proportion of plasmablasts and CD11c+TbethighCD21low B cells (Tbet is also known as TBX21). This subset is known to be autoimmune-prone and displayed even greater autoreactive features in DS including receptors with fewer non-reference nucleotides and higher IGHV4-34 utilization. In vitro, incubation of naive B cells in the plasma of individuals with DS or with IL-6-activated T cells resulted in increased plasmablast differentiation compared with control plasma or unstimulated T cells, respectively. Finally, we detected 365 auto-antibodies in the plasma of individuals with DS, which targeted the gastrointestinal tract, the pancreas, the thyroid, the central nervous system, and the immune system itself. Together, these data point to an autoimmunity-prone state in DS, in which a steady-state cytokinopathy, hyperactivated CD4 T cells and ongoing B cell activation all contribute to a breach in immune tolerance. Our findings also open therapeutic paths, as we demonstrate that T cell activation is resolved not only with broad immunosuppressants such as Jak inhibitors, but also with the more tailored approach of IL-6 inhibition.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
D.B. is the founder and part owner of Lab11 Therapeutics. S.G. reports other research funding from Genentech, Boehringer-Ingelheim, Celgene, Takeda, and Regeneron.
Figures






Comment in
-
Immune dysregulation in Down syndrome.Nat Rev Immunol. 2023 Apr;23(4):201. doi: 10.1038/s41577-023-00855-z. Nat Rev Immunol. 2023. PMID: 36914822 Free PMC article.
References
-
- Ryan, C., Vellody, K., Belazarian, L. & Rork, J. F. Dermatologic conditions in Down syndrome. Pediatr. Dermatol.38, 14731 (2021). - PubMed
-
- Down, J. L. H. Observations on an ethnic classification of idiots. Lond. Hosp. Rep.3, 259–262 (1866).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

