FrangiPANe, a tool for creating a panreference using left behind reads
- PMID: 36814455
- PMCID: PMC9940456
- DOI: 10.1093/nargab/lqad013
FrangiPANe, a tool for creating a panreference using left behind reads
Abstract
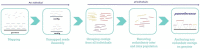
We present here FrangiPANe, a pipeline developed to build panreference using short reads through a map-then-assemble strategy. Applying it to 248 African rice genomes using an improved CG14 reference genome, we identified an average of 8 Mb of new sequences and 5290 new contigs per individual. In total, 1.4 G of new sequences, consisting of 1 306 676 contigs, were assembled. We validated 97.7% of the contigs of the TOG5681 cultivar individual assembly from short reads on a newly long reads genome assembly of the same TOG5681 cultivar. FrangiPANe also allowed the anchoring of 31.5% of the new contigs within the CG14 reference genome, with a 92.5% accuracy at 2 kb span. We annotated in addition 3252 new genes absent from the reference. FrangiPANe was developed as a modular and interactive application to simplify the construction of a panreference using the map-then-assemble approach. It is available as a Docker image containing (i) a Jupyter notebook centralizing codes, documentation and interactive visualization of results, (ii) python scripts and (iii) all the software and libraries requested for each step of the analysis. We foreseen our approach will help leverage large-scale illumina dataset for pangenome studies in GWAS or detection of selection.
© The Author(s) 2023. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures
References
-
- Tranchant-Dubreuil C., Rouard M., Sabot F.. Plant Pangenome: impacts on Phenotypes and Evolution. Annual Plant Reviews online. 2019; 2:453–478.
-
- Bayer P.E., Golicz A.A., Scheben A., Batley J., Edwards D.. Plant pan-genomes are the new reference. Nat. Plants. 2020; 6:914–920. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

