Detection of Pathogens and Antimicrobial Resistance Genes in Ventilator-Associated Pneumonia by Metagenomic Next-Generation Sequencing Approach
- PMID: 36814827
- PMCID: PMC9939671
- DOI: 10.2147/IDR.S397755
Detection of Pathogens and Antimicrobial Resistance Genes in Ventilator-Associated Pneumonia by Metagenomic Next-Generation Sequencing Approach
Abstract
Background: The early identification of pathogens and their antibiotic resistance are essential for the management and treatment of patients affected by ventilator-associated pneumonia (VAP). However, microbiological culture may be time-consuming and has a limited culturability of many potential pathogens. In this study, we developed a rapid nanopore-based metagenomic next-generation sequencing (mNGS) diagnostic assay for detection of VAP pathogens and antimicrobial resistance genes (ARGs).
Patients and methods: Endotracheal aspirate (ETA) samples from 63 patients with suspected VAP were collected between November 2021 and July 2022. Receiver operating characteristic (ROC) curves were established to compare the pathogen identification performance of the target pathogen reads, reads percent of microbes (RPM) and relative abundance (RA). The evaluation of the accuracy of mNGS was performed comparing with the gold standard and the composite standard, respectively. Then, the ARGs were analyzed by mNGS.
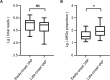
Results: ROC curves showed that RA has the highest diagnostic value and the corresponding threshold was 9.93%. The sensitivity and specificity of mNGS test were 91.3% and 78.3%, respectively, based on the gold standard, while the sensitivity and specificity of mNGS test were 97.4% and 100%, respectively, based on the composite standard. A total of 13 patients were virus-positive based on mNGS results, while the coinfection rate increased from 27% to 46% compared to the rate obtained based on clinical findings. The mNGS test also performed well at predicting antimicrobial resistance phenotypes. Patients with a late-onset VAP had a significantly greater proportion of ARGs in their respiratory microbiome compared to those with early-onset VAP (P = 0.041). Moreover, the median turnaround time of mNGS was 4.43 h, while routine culture was 72.00 h.
Conclusion: In this study, we developed a workflow that can accurately detect VAP pathogens and enable prediction of antimicrobial resistance phenotypes within 5 h of sample receipt by mNGS.
Keywords: NGS; antibiotic resistance; endotracheal aspirate; mechanical ventilation; pathogen diagnosis.
© 2023 Chen et al.
Conflict of interest statement
Ting Chen, Wenhua Huang, Huijun Zong, Qingyu Lv, Yongqiang Jiang, Yan Li, and Peng Liu report grants from National Natural Science Foundation of China, during the conduct of the study. The authors report no conflicts of interest in this work.
Figures





Similar articles
-
Application of mNGS in the Etiological Analysis of Lower Respiratory Tract Infections and the Prediction of Drug Resistance.Microbiol Spectr. 2022 Feb 23;10(1):e0250221. doi: 10.1128/spectrum.02502-21. Epub 2022 Feb 16. Microbiol Spectr. 2022. PMID: 35171007 Free PMC article.
-
Metagenomics for the microbiological diagnosis of hospital-acquired pneumonia and ventilator-associated pneumonia (HAP/VAP) in intensive care unit (ICU): a proof-of-concept study.Respir Res. 2023 Nov 15;24(1):285. doi: 10.1186/s12931-023-02597-x. Respir Res. 2023. PMID: 37968636 Free PMC article.
-
Diagnostic value of metagenomic next-generation sequencing of bronchoalveolar lavage fluid for the diagnosis of suspected pneumonia in immunocompromised patients.BMC Infect Dis. 2022 Apr 29;22(1):416. doi: 10.1186/s12879-022-07381-8. BMC Infect Dis. 2022. PMID: 35488253 Free PMC article.
-
Metagenomics next-generation sequencing tests take the stage in the diagnosis of lower respiratory tract infections.J Adv Res. 2021 Sep 29;38:201-212. doi: 10.1016/j.jare.2021.09.012. eCollection 2022 May. J Adv Res. 2021. PMID: 35572406 Free PMC article. Review.
-
Diagnostic accuracy of the metagenomic next-generation sequencing (mNGS) for detection of bacterial meningoencephalitis: a systematic review and meta-analysis.Eur J Clin Microbiol Infect Dis. 2022 Jun;41(6):881-891. doi: 10.1007/s10096-022-04445-0. Epub 2022 Apr 26. Eur J Clin Microbiol Infect Dis. 2022. PMID: 35474146
Cited by
-
Diagnostic Significance of Targeted Next-Generation Sequencing in Central Nervous System Infections in Neurosurgery of Pediatrics.Infect Drug Resist. 2023 Apr 15;16:2227-2236. doi: 10.2147/IDR.S404277. eCollection 2023. Infect Drug Resist. 2023. PMID: 37090034 Free PMC article.
-
Laboratory tools for the direct detection of bacterial respiratory infections and antimicrobial resistance: a scoping review.J Vet Diagn Invest. 2024 May;36(3):400-417. doi: 10.1177/10406387241235968. Epub 2024 Mar 8. J Vet Diagn Invest. 2024. PMID: 38456288 Free PMC article.
-
Transforming Microbiological Diagnostics in Nosocomial Lower Respiratory Tract Infections: Innovations Shaping the Future.Diagnostics (Basel). 2025 Jan 23;15(3):265. doi: 10.3390/diagnostics15030265. Diagnostics (Basel). 2025. PMID: 39941194 Free PMC article. Review.
-
Effect of metagenomic next-generation sequencing on clinical outcomes in adults with severe pneumonia post-cardiac surgery: a single-center retrospective study.Sci Rep. 2024 Nov 21;14(1):28907. doi: 10.1038/s41598-024-79843-9. Sci Rep. 2024. PMID: 39572618 Free PMC article.
-
Bacterial enrichment prior to third-generation metagenomic sequencing improves detection of BRD pathogens and genetic determinants of antimicrobial resistance in feedlot cattle.Front Microbiol. 2024 May 8;15:1386319. doi: 10.3389/fmicb.2024.1386319. eCollection 2024. Front Microbiol. 2024. PMID: 38779502 Free PMC article.
References
-
- Kalil AC, Metersky ML, Klompas M, et al. Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 Clinical Practice Guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin Infect Dis. 2016;63(5):e61–e111. doi:10.1093/cid/ciw353 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

