Precise and heritable gene targeting in rice using a sequential transformation strategy
- PMID: 36814841
- PMCID: PMC9939429
- DOI: 10.1016/j.crmeth.2022.100389
Precise and heritable gene targeting in rice using a sequential transformation strategy
Abstract
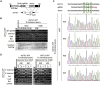
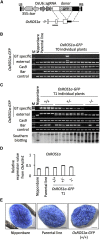
Gene targeting (GT) is a powerful tool for modifying endogenous genomic sequences of interest, such as sequence replacement and gene knockin. Although the efficiency of GT is extremely low in higher plants, engineered sequence-specific nucleases (SSNs)-mediated double-strand breaks (DSBs) can improve GT frequency. We recently reported a CRISPR-Cas9-mediated approach for heritable GT in Arabidopsis, called the "sequential transformation" strategy. For efficient establishment of GT via the sequential transformation method, strong Cas9 activity and robust DSBs are required in the plant cells being infected with Agrobacterium carrying sgRNA and donor DNA. Accordingly, we generated two independent parental lines with maize Ubiquitin 1 promoter-driven Cas9 and established sequential transformation-mediated GT in the Japonica rice cultivar Oryza sativa Nipponbare. We achieved precise GFP knockin into the endogenous OsFTL1 and OsROS1a loci. We believe that our GT technology could be widely utilized in rice research and breeding applications.
Keywords: CRISPR-Cas9; DNA methylation; OsFTL1; OsROS1a; epigenetics; gene targeting; genome editing; genome engineering; rice.
© 2022 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Cas12a-mediated gene targeting by sequential transformation strategy in Arabidopsis thaliana.BMC Plant Biol. 2024 Jul 12;24(1):665. doi: 10.1186/s12870-024-05375-z. BMC Plant Biol. 2024. PMID: 38997669 Free PMC article.
-
Application of multiple sgRNAs boosts efficiency of CRISPR/Cas9-mediated gene targeting in Arabidopsis.BMC Biol. 2024 Jan 17;22(1):6. doi: 10.1186/s12915-024-01810-7. BMC Biol. 2024. PMID: 38233866 Free PMC article.
-
Insights into the molecular mechanisms of CRISPR/Cas9-mediated gene targeting at multiple loci in Arabidopsis.Plant Physiol. 2022 Nov 28;190(4):2203-2216. doi: 10.1093/plphys/kiac431. Plant Physiol. 2022. PMID: 36106983 Free PMC article.
-
Precise Genome Modification via Sequence-Specific Nucleases-Mediated Gene Targeting for Crop Improvement.Front Plant Sci. 2016 Dec 20;7:1928. doi: 10.3389/fpls.2016.01928. eCollection 2016. Front Plant Sci. 2016. PMID: 28066481 Free PMC article. Review.
-
Gene Editing With TALEN and CRISPR/Cas in Rice.Prog Mol Biol Transl Sci. 2017;149:81-98. doi: 10.1016/bs.pmbts.2017.04.006. Epub 2017 May 24. Prog Mol Biol Transl Sci. 2017. PMID: 28712502 Review.
Cited by
-
Double step screening using endogenous marker improves relative gene targeting efficiency in Arabidopsis.Sci Rep. 2024 Dec 28;14(1):30791. doi: 10.1038/s41598-024-80352-y. Sci Rep. 2024. PMID: 39730554 Free PMC article.
-
CRISPR/Cas9: a sustainable technology to enhance climate resilience in major Staple Crops.Front Genome Ed. 2025 Mar 18;7:1533197. doi: 10.3389/fgeed.2025.1533197. eCollection 2025. Front Genome Ed. 2025. PMID: 40171546 Free PMC article. Review.
-
Cas12a-mediated gene targeting by sequential transformation strategy in Arabidopsis thaliana.BMC Plant Biol. 2024 Jul 12;24(1):665. doi: 10.1186/s12870-024-05375-z. BMC Plant Biol. 2024. PMID: 38997669 Free PMC article.
-
Scarless genome editing technology and its application to crop improvement.Breed Sci. 2024 Mar;74(1):32-36. doi: 10.1270/jsbbs.23045. Epub 2024 Mar 9. Breed Sci. 2024. PMID: 39246436 Free PMC article.
-
Application of multiple sgRNAs boosts efficiency of CRISPR/Cas9-mediated gene targeting in Arabidopsis.BMC Biol. 2024 Jan 17;22(1):6. doi: 10.1186/s12915-024-01810-7. BMC Biol. 2024. PMID: 38233866 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

