PARP1pred: a web server for screening the bioactivity of inhibitors against DNA repair enzyme PARP-1
- PMID: 36814851
- PMCID: PMC9939779
- DOI: 10.17179/excli2022-5602
PARP1pred: a web server for screening the bioactivity of inhibitors against DNA repair enzyme PARP-1
Abstract
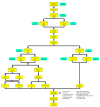
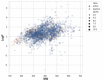
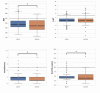
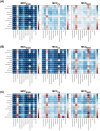
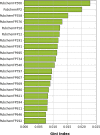
Cancer is the leading cause of death worldwide, resulting in the mortality of more than 10 million people in 2020, according to Global Cancer Statistics 2020. A potential cancer therapy involves targeting the DNA repair process by inhibiting PARP-1. In this study, classification models were constructed using a non-redundant set of 2018 PARP-1 inhibitors. Briefly, compounds were described by 12 fingerprint types and built using the random forest algorithm concomitant with various sampling approaches. Results indicated that PubChem with an oversampling approach yielded the best performance, with a Matthews correlation coefficient > 0.7 while also affording interpretable molecular features. Moreover, feature importance, as determined from the Gini index, revealed that the aromatic/cyclic/heterocyclic moiety, nitrogen-containing fingerprints, and the ether/aldehyde/alcohol moiety were important for PARP-1 inhibition. Finally, our predictive model was deployed as a web application called PARP1pred and is publicly available at https://parp1pred.streamlitapp.com, allowing users to predict the biological activity of query compounds using their SMILES notation as the input. It is anticipated that the model described herein will aid in the discovery of effective PARP-1 inhibitors.
Keywords: DNA repair; PARP-1; QSAR; cheminformatics; machine learning; webserver.
Copyright © 2023 Lerksuthirat et al.
Figures




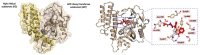
References
-
- Abbasi-Radmoghaddam Z, Riahi S, Gharaghani S, Mohammadi-Khanaposhtanai M. Design of potential anti-tumor PARP-1 inhibitors by QSAR and molecular modeling studies. Mol Diversity. 2021;25:263–277. - PubMed
-
- Balasubramaniam S, Beaver JA, Horton S, Fernandes LL, Tang S, Horne HN, et al. fda approval summary: Rucaparib for the treatment of patients with deleterious BRCA mutation-associated advanced ovarian cancer. Clin Cancer Res. 2017;23:7165–7170. - PubMed
-
- Banasik M, Komura H, Shimoyama M, Ueda K. Specific inhibitors of poly(ADP-ribose) synthetase and mono(ADP-ribosyl)transferase. J Biol Chem. 1992;267:1569–1575. - PubMed
-
- Baudino TA. Targeted cancer therapy: the next generation of cancer treatment. Curr Drug Discov Technol. 2015;12:3–20. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
