Genome-wide association study reveals genetic loci and candidate genes for meat quality traits in a four-way crossbred pig population
- PMID: 36814900
- PMCID: PMC9939654
- DOI: 10.3389/fgene.2023.1001352
Genome-wide association study reveals genetic loci and candidate genes for meat quality traits in a four-way crossbred pig population
Abstract
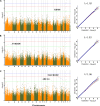
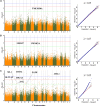
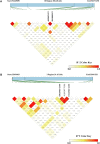
Meat quality traits (MQTs) have gained more attention from breeders due to their increasing economic value in the commercial pig industry. In this genome-wide association study (GWAS), 223 four-way intercross pigs were genotyped using the specific-locus amplified fragment sequencing (SLAF-seq) and phenotyped for PH at 45 min post mortem (PH45), meat color score (MC), marbling score (MA), water loss rate (WL), drip loss (DL) in the longissimus muscle, and cooking loss (CL) in the psoas major muscle. A total of 227, 921 filtered single nucleotide polymorphisms (SNPs) evenly distributed across the entire genome were detected to perform GWAS. A total of 64 SNPs were identified for six meat quality traits using the mixed linear model (MLM), of which 24 SNPs were located in previously reported QTL regions. The phenotypic variation explained (PVE) by the significant SNPs was from 2.43% to 16.32%. The genomic heritability estimates based on SNP for six meat-quality traits were low to moderate (0.07-0.47) being the lowest for CL and the highest for DL. A total of 30 genes located within 10 kb upstream or downstream of these significant SNPs were found. Furthermore, several candidate genes for MQTs were detected, including pH45 (GRM8), MC (ANKRD6), MA (MACROD2 and ABCG1), WL (TMEM50A), CL (PIP4K2A) and DL (CDYL2, CHL1, ABCA4, ZAG and SLC1A2). This study provided substantial new evidence for several candidate genes to participate in different pork quality traits. The identification of these SNPs and candidate genes provided a basis for molecular marker-assisted breeding and improvement of pork quality traits.
Keywords: GWAS; SLAF-seq; candidate genes; crossbred pigs; meat quality.
Copyright © 2023 Wang, Wang, Li, Sun, Chen, Yan, Dong, Pan and Lu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Genome-wide identification of quantitative trait loci and candidate genes for seven carcass traits in a four-way intercross porcine population.BMC Genomics. 2024 Jun 10;25(1):582. doi: 10.1186/s12864-024-10484-y. BMC Genomics. 2024. PMID: 38858624 Free PMC article.
-
Genome-wide association study identifying genetic variants associated with carcass backfat thickness, lean percentage and fat percentage in a four-way crossbred pig population using SLAF-seq technology.BMC Genomics. 2022 Aug 15;23(1):594. doi: 10.1186/s12864-022-08827-8. BMC Genomics. 2022. PMID: 35971078 Free PMC article.
-
Genome-Wide Association Study of Growth Traits in a Four-Way Crossbred Pig Population.Genes (Basel). 2022 Oct 31;13(11):1990. doi: 10.3390/genes13111990. Genes (Basel). 2022. PMID: 36360227 Free PMC article.
-
The Genetic Architecture of Meat Quality Traits in a Crossbred Commercial Pig Population.Foods. 2022 Oct 9;11(19):3143. doi: 10.3390/foods11193143. Foods. 2022. PMID: 36230219 Free PMC article.
-
Genome-Wide Association Study of Meat Quality Traits in a Three-Way Crossbred Commercial Pig Population.Front Genet. 2021 Mar 17;12:614087. doi: 10.3389/fgene.2021.614087. eCollection 2021. Front Genet. 2021. PMID: 33815461 Free PMC article.
Cited by
-
Integration of transcriptome and machine learning to identify the potential key genes and regulatory networks affecting drip loss in pork.J Anim Sci. 2024 Jan 3;102:skae164. doi: 10.1093/jas/skae164. J Anim Sci. 2024. PMID: 38865489 Free PMC article.
-
Genome-Wide Association Studies of Hair Whorl in Pigs.Genes (Basel). 2024 Sep 25;15(10):1249. doi: 10.3390/genes15101249. Genes (Basel). 2024. PMID: 39457372 Free PMC article.
-
Application of GWAS and mGWAS in Livestock and Poultry Breeding.Animals (Basel). 2024 Aug 16;14(16):2382. doi: 10.3390/ani14162382. Animals (Basel). 2024. PMID: 39199916 Free PMC article. Review.
-
Genome-wide identification of quantitative trait loci and candidate genes for seven carcass traits in a four-way intercross porcine population.BMC Genomics. 2024 Jun 10;25(1):582. doi: 10.1186/s12864-024-10484-y. BMC Genomics. 2024. PMID: 38858624 Free PMC article.
-
Deciphering the Population Characteristics of Leiqiong Cattle Using Whole-Genome Sequencing Data.Animals (Basel). 2025 Jan 24;15(3):342. doi: 10.3390/ani15030342. Animals (Basel). 2025. PMID: 39943110 Free PMC article.
References
-
- Aaslyng M. D., Bejerholma C., Ertbjergb P., Bertramc H. C., Andersenc H. J. (2003). Cooking loss and juiciness of pork in relation to raw meat quality and cooking procedure. Food Qual. Prefer. 14, 277–288. 10.1016/s0950-3293(02)00086-1 - DOI
-
- Carricaburu V., Lamia K. A., Lo E., Favereaux L., Payrastre B., Cantley L. C., et al. (2003). The phosphatidylinositol (PI)-5-phosphate 4-kinase type II enzyme controls insulin signaling by regulating PI-3,4,5-trisphosphate degradation. Proc. Natl. Acad. Sci. U. S. A. 100, 9867–9872. 10.1073/pnas.1734038100 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

