Differential expression and effect analysis of lncRNA-mRNA in congenital pseudarthrosis of the tibia
- PMID: 36814904
- PMCID: PMC9939773
- DOI: 10.3389/fgene.2023.1094298
Differential expression and effect analysis of lncRNA-mRNA in congenital pseudarthrosis of the tibia
Abstract
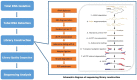
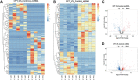
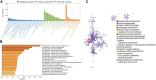
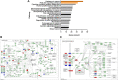
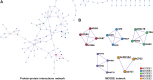
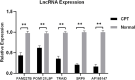
Background: To analyze the lncRNA-mRNA differential expression and co-expression network of periosteal stem cells (PSCs) from congenital pseudarthrosis of the tibia (CPT) and normal patients, and to explore the role of key lncRNAs. Methods: Differentially expressed lncRNAs and mRNAs in PSCs were obtained by sequencing, and biological functions of differentially expressed mRNAs were detected by gene ontology (GO), Kyoto encyclopedia of genes and genomes (KEGG) pathway and protein -protein interaction (PPI) analysis. The co-expression network of lncRNA-mRNA was constructed by correlation analysis of differentially expressed lncRNAs and mRNAs, and the key lncRNAs were screened according to the connectivity degree. After that, the cis-regulated target genes of differential expressed lncRNAs and mRNAs were predicted. Results: A total of 194 differentially expressed lncRNAs were identified, including 73 upregulated and 121 downregulated genes. A total of 822 differentially expressed mRNAs were identified, including 311 upregulated and 511 downregulated genes. GO, KEGG and PPI enrichment analysis showed that the regulatory function of differentially expressed mRNAs were mainly gathered in skeletal system development and tissue morphogenesis. The co-expression network with 226 nodes and 3,390 edges was constructed based on correlation analysis. A total of 10 key lncRNAs, including FAM227B, POM121L9P, AF165147 and AC103702, were screened according to connectivity degree. Prediction of target genes indicated that FAM227B-FGF7 and AC103702-HOXB4/5/6 may play an important role in the pathogenesis of CPT. Conclusion: A total of 10 key lncRNAs, including FAM227B, POM121L9P, AF165147, and AC103702, occupy the core position in the co-expression network, suggesting that these lncRNAs and their target genes may play an important role in the pathogenesis of CPT.
Keywords: bioinformatic analysis; congenital pseudarthrosis of the tibia; long non-coding RNA; messenger RNA; periosteal stem cell.
Copyright © 2023 Li, Mei, Liu and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Identification of key genes for hypertrophic cardiomyopathy using integrated network analysis of differential lncRNA and gene expression.Front Cardiovasc Med. 2022 Aug 4;9:946229. doi: 10.3389/fcvm.2022.946229. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 35990977 Free PMC article.
-
Comprehensive analysis of lncRNA-miRNA-mRNA networks during osteogenic differentiation of bone marrow mesenchymal stem cells.BMC Genomics. 2022 Jun 7;23(1):425. doi: 10.1186/s12864-022-08646-x. BMC Genomics. 2022. PMID: 35672672 Free PMC article.
-
Characterization of differentially expressed and lipid metabolism-related lncRNA-mRNA interaction networks during the growth of liver tissue through rabbit models.Front Vet Sci. 2022 Sep 1;9:998796. doi: 10.3389/fvets.2022.998796. eCollection 2022. Front Vet Sci. 2022. PMID: 36118359 Free PMC article.
-
Integrated analysis of immune- and apoptosis-related lncRNA-miRNA-mRNA regulatory network in children with Henoch Schönlein purpura nephritis.Transl Pediatr. 2022 Oct;11(10):1682-1696. doi: 10.21037/tp-22-437. Transl Pediatr. 2022. PMID: 36345450 Free PMC article.
-
Genome-Wide Analysis of Differentially Expressed mRNAs and lncRNAs in Koi Carp Infected with Koi Herpesvirus.Viruses. 2022 Nov 18;14(11):2555. doi: 10.3390/v14112555. Viruses. 2022. PMID: 36423164 Free PMC article.
Cited by
-
Circ_0000888 regulates osteogenic differentiation of periosteal mesenchymal stem cells in congenital pseudarthrosis of the tibia.iScience. 2023 Sep 14;26(10):107923. doi: 10.1016/j.isci.2023.107923. eCollection 2023 Oct 20. iScience. 2023. PMID: 37810257 Free PMC article.
-
Long non-coding RNA SNHG4 aggravates cigarette smoke-induced COPD by regulating miR-144-3p/EZH2 axis.BMC Pulm Med. 2023 Dec 19;23(1):513. doi: 10.1186/s12890-023-02818-5. BMC Pulm Med. 2023. PMID: 38114929 Free PMC article.
-
Unraveling the molecular landscape of congenital pseudoarthrosis of the tibia: insights from a comprehensive analysis of 159 probands.Orphanet J Rare Dis. 2025 Jun 3;20(1):269. doi: 10.1186/s13023-025-03759-4. Orphanet J Rare Dis. 2025. PMID: 40462134 Free PMC article.
References
LinkOut - more resources
Full Text Sources

