Genetic architecture of innate and adaptive immune cells in pigs
- PMID: 36814923
- PMCID: PMC9939681
- DOI: 10.3389/fimmu.2023.1058346
Genetic architecture of innate and adaptive immune cells in pigs
Abstract
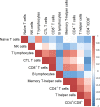
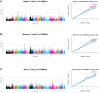
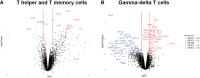
Pig industry is facing new challenges that make necessary to reorient breeding programs to produce more robust and resilient pig populations. The aim of the present work was to study the genetic determinism of lymphocyte subpopulations in the peripheral blood of pigs and identify genomic regions and biomarkers associated to them. For this purpose, we stained peripheral blood mononuclear cells to measure ten immune-cell-related traits including the relative abundance of different populations of lymphocytes, the proportions of CD4+ T cells and CD8+ T cells, and the ratio of CD4+/CD8+ T cells from 391 healthy Duroc piglets aged 8 weeks. Medium to high heritabilities were observed for the ten immune-cell-related traits and significant genetic correlations were obtained between the proportion of some lymphocytes populations. A genome-wide association study pointed out 32 SNPs located at four chromosomal regions on pig chromosomes SSC3, SSC5, SSC8, and SSCX as significantly associated to T-helper cells, memory T-helper cells and γδ T cells. Several genes previously identified in human association studies for the same or related traits were located in the associated regions, and were proposed as candidate genes to explain the variation of T cell populations such as CD4, CD8A, CD8B, KLRC2, RMND5A and VPS24. The transcriptome analysis of whole blood samples from animals with extreme proportions of γδ T, T-helper and memory T-helper cells identified differentially expressed genes (CAPG, TCF7L1, KLRD1 and CD4) located into the associated regions. In addition, differentially expressed genes specific of different T cells subpopulations were identified such as SOX13 and WC1 genes for γδ T cells. Our results enhance the knowledge about the genetic control of lymphocyte traits that could be considered to optimize the induction of immune responses to vaccines against pathogens. Furthermore, they open the possibility of applying effective selection programs for improving immunocompetence in pigs and support the use of the pig as a very reliable human biomedical model.
Keywords: RNA-Seq; biomodel; immune cells; immunocompentence; pig; γδ T cells.
Copyright © 2023 Ballester, Jové-Juncà, Pascual, López-Serrano, Crespo-Piazuelo, Hernández-Banqué, González-Rodríguez, Ramayo-Caldas and Quintanilla.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Genetic parameters and associated genomic regions for global immunocompetence and other health-related traits in pigs.Sci Rep. 2020 Oct 28;10(1):18462. doi: 10.1038/s41598-020-75417-7. Sci Rep. 2020. PMID: 33116177 Free PMC article.
-
Genome-wide association study for T lymphocyte subpopulations in swine.BMC Genomics. 2012 Sep 18;13:488. doi: 10.1186/1471-2164-13-488. BMC Genomics. 2012. PMID: 22985182 Free PMC article.
-
Discrimination of different subsets of cytolytic cells in pseudorabies virus-immune and naive pigs.J Gen Virol. 2000 Jun;81(Pt 6):1529-37. doi: 10.1099/0022-1317-81-6-1529. J Gen Virol. 2000. PMID: 10811936
-
Porcine gammadelta T cells: possible roles on the innate and adaptive immune responses following virus infection.Vet Immunol Immunopathol. 2006 Jul 15;112(1-2):49-61. doi: 10.1016/j.vetimm.2006.03.011. Epub 2006 May 22. Vet Immunol Immunopathol. 2006. PMID: 16714063 Review.
-
Characterization of porcine T lymphocytes and their immune response against viral antigens.J Biotechnol. 1999 Aug 20;73(2-3):223-33. doi: 10.1016/s0168-1656(99)00140-6. J Biotechnol. 1999. PMID: 10486931 Review.
Cited by
-
Unveiling regulatory variants in the blood transcriptome and their association with immunity traits in pigs.Front Immunol. 2025 Jun 5;16:1582982. doi: 10.3389/fimmu.2025.1582982. eCollection 2025. Front Immunol. 2025. PMID: 40539042 Free PMC article.
-
Identification of SNPs and Candidate Genes Associated with Monocyte/Lymphocyte Ratio and Neutrophil/Lymphocyte Ratio in Duroc × Erhualian F2 Population.Int J Mol Sci. 2024 Sep 9;25(17):9745. doi: 10.3390/ijms25179745. Int J Mol Sci. 2024. PMID: 39273692 Free PMC article.
-
Pig jejunal single-cell RNA landscapes revealing breed-specific immunology differentiation at various domestication stages.Front Immunol. 2025 Feb 28;16:1530214. doi: 10.3389/fimmu.2025.1530214. eCollection 2025. Front Immunol. 2025. PMID: 40151618 Free PMC article.
-
Integrating large-scale meta-GWAS and PigGTEx resources to decipher the genetic basis of 232 complex traits in pigs.Natl Sci Rev. 2025 Feb 17;12(5):nwaf048. doi: 10.1093/nsr/nwaf048. eCollection 2025 May. Natl Sci Rev. 2025. PMID: 40330097 Free PMC article.
-
Insights into genetic determinants of piglet survival during a PRRSV outbreak.Vet Res. 2024 Dec 18;55(1):160. doi: 10.1186/s13567-024-01421-8. Vet Res. 2024. PMID: 39696499 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

