H3K4 Methylation Promotes Expression of Mitochondrial Dynamics Regulators to Ensure Oocyte Quality in Mice
- PMID: 36815388
- PMCID: PMC10131798
- DOI: 10.1002/advs.202204794
H3K4 Methylation Promotes Expression of Mitochondrial Dynamics Regulators to Ensure Oocyte Quality in Mice
Abstract
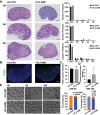
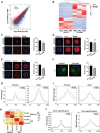
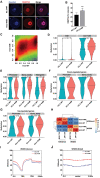
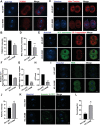
Significantly decreased H3K4 methylation in oocytes from aged mice indicates the important roles of H3K4 methylation in female reproduction. However, how H3K4 methylation regulates oocyte development remains largely unexplored. In this study, it is demonstrated that oocyte-specific expression of dominant negative mutant H3.3-K4M led to a decrease of the level of H3K4 methylation in mouse oocytes, resulting in reduced transcriptional activity and increased DNA methylation in oocytes, disturbed oocyte developmental potency, and fertility of female mice. The impaired expression of genes regulating mitochondrial functions in H3.3-K4M oocytes, accompanied by mitochondrial abnormalities, is further noticed. Moreover, early embryos from H3.3-K4M oocytes show developmental arrest and reduced zygotic genome activation. Collectively, these results show that H3K4 methylation in oocytes is critical to orchestrating gene expression profile, driving the oocyte developmental program, and ensuring oocyte quality. This study also improves understanding of how histone modifications regulate organelle dynamics in oocytes.
Keywords: H3K4 methylation; mitochondrial dysfunction; oocyte; oogenesis; zygotic genome activation.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Vander Borght M., Wyns C., Clin. Biochem. 2018, 62, 2. - PubMed
-
- Zhou Z., Zheng D., Wu H., Li R., Xu S., Kang Y., Cao Y., Chen X., Zhu Y., Xu S., Chen Z.‐J., Mol B.w, Qiao J., BJOG 2018, 125, 432. - PubMed
-
- Zheng D., Zhou Z., Li R., Wu H., Xu S., Kang Y., Cao Y., Chen X., Zhu Y., Xu S., Chen Z.i‐J., Mol B. W., Qiao J., Reprod. Biomed. Online 2019, 38, 917. - PubMed
-
- Bilmez Y., Talibova G., Ozturk S., Histochem. Cell Biol. 2022, 157, 7. - PubMed
-
- Wang Y., Liu Q., Tang F., Yan L., Qiao J., Annu. Rev. Genomics Hum. Genet. 2019, 20, 21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
