Overexpression of 18S rRNA methyltransferase CrBUD23 enhances biomass and lutein content in Chlamydomonas reinhardtii
- PMID: 36815903
- PMCID: PMC9935685
- DOI: 10.3389/fbioe.2023.1102098
Overexpression of 18S rRNA methyltransferase CrBUD23 enhances biomass and lutein content in Chlamydomonas reinhardtii
Abstract
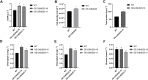
Post-transcriptional modification of nucleic acids including transfer RNA (tRNA), ribosomal RNA (rRNA) and messenger RNA (mRNA) is vital for fine-tunning of mRNA translation. Methylation is one of the most widespread post-transcriptional modifications in both eukaryotes and prokaryotes. HsWBSCR22 and ScBUD23 encodes a 18S rRNA methyltransferase that positively regulates cell growth by mediating ribosome maturation in human and yeast, respectively. However, presence and function of 18S rRNA methyltransferase in green algae are still elusive. Here, through bioinformatic analysis, we identified CrBUD23 as the human WBSCR22 homolog in genome of the green algae model organism Chlamydonomas reinhardtii. CrBUD23 was a conserved putative 18S rRNA methyltransferase widely exited in algae, plants, insects and mammalians. Transcription of CrBUD23 was upregulated by high light and down-regulated by low light, indicating its role in photosynthesis and energy metabolism. To characterize its biological function, coding sequence of CrBUD23 fused with a green fluorescence protein (GFP) tag was derived by 35S promoter and stably integrated into Chlamydomonas genome by glass bead-mediated transformation. Compared to C. reinhardtii wild type CC-5325, transgenic strains overexpressing CrBUD23 resulted in accelerated cell growth, thereby leading to elevated biomass, dry weight and protein content. Moreover, overexpression of CrBUD23 increased content of photosynthetic pigments but not elicit the activation of antioxidative enzymes, suggesting CrBUD23 favors growth and proliferation in the trade-off with stress responses. Bioinformatic analysis revealed the G1177 was the putative methylation site in 18S rRNA of C. reinhardtii CC-849. G1177 was conserved in other Chlamydonomas isolates, indicating the conserved methyltransferase activity of BUD23 proteins. In addition, CrTrm122, the homolog of BUD23 interactor Trm112, was found involved in responses to high light as same as CrBUD23. Taken together, our study revealed that cell growth, protein content and lutein accumulation of Chlamydomonas were positively regulated by the 18S rRNA methyltransferase CrBUD23, which could serve as a promising candidate for microalgae genetic engineering.
Keywords: 18S rRNA methyltransferase; Chlamydomonas reinhardtii; CrBUD23; CrTrm112; biomass; lutein; overexpression.
Copyright © 2023 Liu, Guo, Zhao, Zou, Sun, Feng, Zeng, Wen, Chen, Hu, Lou and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

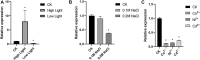





Similar articles
-
Trm112 is required for Bud23-mediated methylation of the 18S rRNA at position G1575.Mol Cell Biol. 2012 Jun;32(12):2254-67. doi: 10.1128/MCB.06623-11. Epub 2012 Apr 9. Mol Cell Biol. 2012. PMID: 22493060 Free PMC article.
-
The human 18S rRNA base methyltransferases DIMT1L and WBSCR22-TRMT112 but not rRNA modification are required for ribosome biogenesis.Mol Biol Cell. 2015 Jun 1;26(11):2080-95. doi: 10.1091/mbc.E15-02-0073. Epub 2015 Apr 7. Mol Biol Cell. 2015. PMID: 25851604 Free PMC article.
-
Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes.Proc Natl Acad Sci U S A. 2014 Dec 23;111(51):E5518-26. doi: 10.1073/pnas.1413089111. Epub 2014 Dec 8. Proc Natl Acad Sci U S A. 2014. PMID: 25489090 Free PMC article.
-
Trm112, a Protein Activator of Methyltransferases Modifying Actors of the Eukaryotic Translational Apparatus.Biomolecules. 2017 Jan 27;7(1):7. doi: 10.3390/biom7010007. Biomolecules. 2017. PMID: 28134793 Free PMC article. Review.
-
Nitrogen-dependent coordination of cell cycle, quiescence and TAG accumulation in Chlamydomonas.Biotechnol Biofuels. 2019 Dec 23;12:292. doi: 10.1186/s13068-019-1635-0. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 31890020 Free PMC article. Review.
Cited by
-
Bioremoval of Co(II) by a novel halotolerant microalgae Dunaliella sp. FACHB-558 from saltwater.Front Microbiol. 2024 Apr 30;15:1256814. doi: 10.3389/fmicb.2024.1256814. eCollection 2024. Front Microbiol. 2024. PMID: 38746752 Free PMC article.
-
Elevated accumulation of lutein and zeaxanthin in a novel high-biomass yielding strain Dunaliella sp. ZP-1 obtained through EMS mutagenesis.Biotechnol Biofuels Bioprod. 2025 Mar 27;18(1):39. doi: 10.1186/s13068-025-02629-2. Biotechnol Biofuels Bioprod. 2025. PMID: 40148979 Free PMC article.
-
Mechanistic insight for improving butenyl-spinosyn production through combined ARTP/UV mutagenesis and ribosome engineering in Saccharopolyspora pogona.Front Bioeng Biotechnol. 2024 Jan 15;11:1329859. doi: 10.3389/fbioe.2023.1329859. eCollection 2023. Front Bioeng Biotechnol. 2024. PMID: 38292303 Free PMC article.
-
Mutagenesis selection and large-scale cultivation of non-green Chlamydomonas reinhardtii for food applications.Front Nutr. 2024 Sep 25;11:1456230. doi: 10.3389/fnut.2024.1456230. eCollection 2024. Front Nutr. 2024. PMID: 39385786 Free PMC article.
-
Nanomedicines Targeting Metabolic Pathways in the Tumor Microenvironment: Future Perspectives and the Role of AI.Metabolites. 2025 Mar 13;15(3):201. doi: 10.3390/metabo15030201. Metabolites. 2025. PMID: 40137165 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

