Genomic epidemiology of Mycobacterium avium subsp. paratuberculosis isolates from Canadian dairy herds provides evidence for multiple infection events
- PMID: 36816022
- PMCID: PMC9934062
- DOI: 10.3389/fgene.2023.1043598
Genomic epidemiology of Mycobacterium avium subsp. paratuberculosis isolates from Canadian dairy herds provides evidence for multiple infection events
Abstract
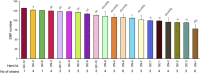
Mycobacterium avium subsp. paratuberculosis (MAP) is the pathogen responsible for paratuberculosis or Johne's Disease (JD) in ruminants, which is responsible for substantial economic losses worldwide. MAP transmission primarily occurs through the fecal-oral route, and the introduction of an MAP infected animal into a herd is an important transmission route. In the current study, we characterized MAP isolates from 67 cows identified in 20 herds from the provinces of Quebec and Ontario, Canada. Whole genome sequencing (WGS) was performed and an average genome coverage (relative to K-10) of ∼14.9 fold was achieved. The total number of SNPs present in each isolate varied from 51 to 132 and differed significantly between herds. Isolates with the highest genetic variability were generally present in herds from Quebec. The isolates were broadly separated into two main clades and this distinction was not influenced by the province from which they originated. Analysis of 8 MIRU-VNTR loci and 11 SSR loci was performed on the 67 isolates from the 20 dairy herds and publicly available references, notably major genetic lineages and six isolates from the province of Newfoundland and Labrador. All 67 field isolates were phylogenetically classified as Type II (C-type) and according to MIRU-VNTR, the predominant type was INMV 2 (76.1%) among four distinct patterns. Multilocus SSR typing identified 49 distinct INMV SSR patterns. The discriminatory index of the multilocus SSR typing was 0.9846, which was much higher than MIRU-VNTR typing (0.3740). Although multilocus SSR analysis provides good discriminatory power, the resolution was not informative enough to determine inter-herd transmission. In select cases, SNP-based analysis was the only approach able to document disease transmission between herds, further validated by animal movement data. The presence of SNPs in several virulence genes, notably for PE, PPE, mce and mmpL, is expected to explain differential antigenic or pathogenetic host responses. SNP-based studies will provide insight into how MAP genetic variation may impact host-pathogen interactions. Our study highlights the informative power of WGS which is now recommended for epidemiological studies and to document mixed genotypes infections.
Keywords: John’s disease; MIRU-VNTR analysis; MLSSR typing; molecular epidemiology; mycobacterium avium subsp. paratuberculosis; phylogenetic SNP based analysis; strain typing; whole genome sequencing (WGS).
Copyright © 2023 Byrne, Ollier, Tahlan, Biet and Bissonnette.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Agrawal G., Aitken J., Hamblin H., Collins M., Borody T. J. (2021). Putting crohn's on the MAP: Five common questions on the contribution of Mycobacterium avium subspecies paratuberculosis to the pathophysiology of crohn's disease. Dig. Dis. Sci. 66, 348–358. 10.1007/s10620-020-06653-0 - DOI - PMC - PubMed
-
- Ahlstrom C., Barkema H. W., De Buck J. (2014). Improved short-sequence-repeat genotyping of Mycobacterium avium subsp. paratuberculosis by using matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 80, 534–539. 10.1128/AEM.03212-13 - DOI - PMC - PubMed
-
- Ahlstrom C., Barkema H. W., Stevenson K., Zadoks R. N., Biek R., Kao R., et al. (2015). Limitations of variable number of tandem repeat typing identified through whole genome sequencing of Mycobacterium avium subsp. paratuberculosis on a national and herd level. BMC Genomics 16, 161. 10.1186/s12864-015-1387-6 - DOI - PMC - PubMed
-
- Amonsin A., Li L. L., Zhang Q., Bannantine J. P., Motiwala A. S., Sreevatsan S., et al. (2004). Multilocus short sequence repeat sequencing approach for differentiating among Mycobacterium avium subsp. paratuberculosis strains. J. Clin. Microbiol. 42, 1694–1702. 10.1128/jcm.42.4.1694-1702.2004 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

