Identification of immune related gene signature for predicting prognosis of cholangiocarcinoma patients
- PMID: 36817485
- PMCID: PMC9932535
- DOI: 10.3389/fimmu.2023.1028404
Identification of immune related gene signature for predicting prognosis of cholangiocarcinoma patients
Abstract
Objective: To identify the gene subtypes related to immune cells of cholangiocarcinoma and construct an immune score model to predict the immunotherapy efficacy and prognosis for cholangiocarcinoma.
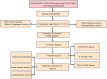
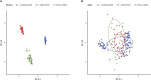
Methods: Based on principal component analysis (PCA) algorithm, The Cancer Genome Atlas (TCGA)-cholangiocarcinoma, GSE107943 and E-MTAB-6389 datasets were combined as Joint data. Immune genes were downloaded from ImmPort. Univariate Cox survival analysis filtered prognostically associated immune genes, which would identify immune-related subtypes of cholangiocarcinoma. Least absolute shrinkage and selection operator (LASSO) further screened immune genes with prognosis values, and tumor immune score was calculated for patients with cholangiocarcinoma after the combination of the three datasets. Kaplan-Meier curve analysis determined the optimal cut-off value, which was applied for dividing cholangiocarcinoma patients into low and high immune score group. To explore the differences in tumor microenvironment and immunotherapy between immune cell-related subtypes and immune score groups of cholangiocarcinoma.
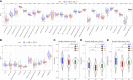
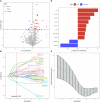
Results: 34 prognostic immune genes and three immunocell-related subtypes with statistically significant prognosis (IC1, IC2 and IC3) were identified. Among them, IC1 and IC3 showed higher immune cell infiltration, and IC3 may be more suitable for immunotherapy and chemotherapy. 10 immune genes with prognostic significance were screened by LASSO regression analysis, and a tumor immune score model was constructed. Kaplan-Meier (KM) and receiver operating characteristic (ROC) analysis showed that RiskScore had excellent prognostic prediction ability. Immunohistochemical analysis showed that 6 gene (NLRX1, AKT1, CSRP1, LEP, MUC4 and SEMA4B) of 10 genes were abnormal expressions between cancer and paracancer tissue. Immune cells infiltration in high immune score group was generally increased, and it was more suitable for chemotherapy. In GSE112366-Crohn's disease dataset, 6 of 10 immune genes had expression differences between Crohn's disease and healthy control. The area under ROC obtained 0.671 based on 10-immune gene signature. Moreover, the model had a sound performance in Crohn's disease.
Conclusion: The prediction of tumor immune score model in predicting immune microenvironment, immunotherapy and chemotherapy in patients with cholangiocarcinoma has shown its potential for indicating the effect of immunotherapy on patients with cholangiocarcinoma.
Keywords: RiskScore; cholangiocarcinoma; immune; immunotherapy; molecular subtype.
Copyright © 2023 Zhang, Huang, Liu, Wang, Zhou, Hou, Tang, Xiong, Wen and Huang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

