After virus exposure, early bystander naïve CD8 T cell activation relies on NAD+ salvage metabolism
- PMID: 36818473
- PMCID: PMC9932030
- DOI: 10.3389/fimmu.2022.1047661
After virus exposure, early bystander naïve CD8 T cell activation relies on NAD+ salvage metabolism
Abstract
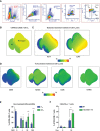
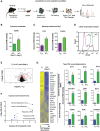
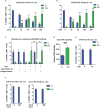
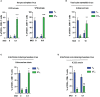
CD8 T cells play a central role in antiviral immunity. Type I interferons are among the earliest responders after virus exposure and can cause extensive reprogramming and antigen-independent bystander activation of CD8 T cells. Although bystander activation of pre-existing memory CD8 T cells is known to play an important role in host defense and immunopathology, its impact on naïve CD8 T cells remains underappreciated. Here we report that exposure to reovirus, both in vitro or in vivo, promotes bystander activation of naïve CD8 T cells within 24 hours and that this distinct subtype of CD8 T cell displays an innate, antiviral, type I interferon sensitized signature. The induction of bystander naïve CD8 T cells is STAT1 dependent and regulated through nicotinamide phosphoribosyl transferase (NAMPT)-mediated enzymatic actions within NAD+ salvage metabolic biosynthesis. These findings identify a novel aspect of CD8 T cell activation following virus infection with implications for human health and physiology.
Keywords: CD8 T cells; NAD+ salvage metabolism; antiviral immunity; bystander activation; immunometabolism; metabolic reprogramming; naïve CD8 T cells; type I interferons.
Copyright © 2023 Holay, Kennedy, Murphy, Konda, Giacomantonio, Brauer-Chapin, Paulo, Kumar, Kim, Elaghil, Sisson, Clements, Richardson, Gygi and Gujar.
Conflict of interest statement
Authors BK and ME were employed by company IMV Inc,. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Snell LM, MacLeod BL, Law JC, Osokine I, Elsaesser HJ, Hezaveh K, et al. . CD8+ T cell priming in established chronic viral infection preferentially directs differentiation of memory-like cells for sustained immunity. Immunity (2018) 49:678–694.e5. doi: 10.1016/j.immuni.2018.08.002 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

