An immune-suppressing protein in human endogenous retroviruses
- PMID: 36818731
- PMCID: PMC9927554
- DOI: 10.1093/bioadv/vbad013
An immune-suppressing protein in human endogenous retroviruses
Abstract
Motivation: Retroviruses are important contributors to disease and evolution in vertebrates. Sometimes, retrovirus DNA is heritably inserted in a vertebrate genome: an endogenous retrovirus (ERV). Vertebrate genomes have many such virus-derived fragments, usually with mutations disabling their original functions.
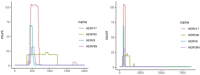
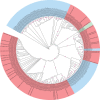
Results: Some primate ERVs appear to encode an overlooked protein. This protein is homologous to protein MC132 from Molluscum contagiosum virus, which is a human poxvirus, not a retrovirus. MC132 suppresses the immune system by targeting NF- B, and it had no known homologs until now. The ERV homologs of MC132 in the human genome are mostly disrupted by mutations, but there is an intact copy on chromosome 4. We found homologs of MC132 in ERVs of apes, monkeys and bushbaby, but not tarsiers, lemurs or non-primates. This suggests that some primate retroviruses had, or have, an extra immune-suppressing protein, which underwent horizontal genetic transfer between unrelated viruses.
Contact: mcfrith@edu.k.u-tokyo.ac.jp.
© The Author(s) 2023. Published by Oxford University Press.
Figures





References
LinkOut - more resources
Full Text Sources

